Nasopharyngeal carcinoma (NPC) starts from the squamous epithelium of the nasopharynx; its etiology is multifactorial and implicates Epstein–Barr virus infection, genetic predispositions, and environmental elements such as alcohol and tobacco use.[1] This malignancy predominantly affects populations in East and Southeast Asia, and the vast majority of diagnoses present as locally advanced stages.[2] The initial management for NPC frequently involves platinum-based concurrent chemo-radiotherapy;[3] however, high-dose cisplatin administration frequently encounters resistance, which not only impedes therapeutic success but also escalates recurrence rates and metastatic potential, and poses a critical barrier to effective NPC therapy.[4] Emerging research highlights the significant role of extracellular vesicles in fostering chemo-resistance within cancer cells and signifies their potential utility as diagnostic markers and therapeutic targets in oncology.[5] Exosomes are membrane vesicles composed of a phospholipid bilayer that can be secreted by a wide range of cells, including cancer cells, and contain diverse proteins, lipids, and nucleic acids.[6] When exosomes fuse with the plasma membrane, they are released into the extracellular environment, where they play a role in altering the tumor micro- environment (TME) by transferring macro molecules.[7] Exosomes deliver cellular molecular components (including nucleic acids and proteins) that modify the physiological state and biological functions of recipient cells, and thereby promote cell growth, metastasis, angiogenesis, and participation in radiotherapy resistance;[8] therefore, clarifying the possible mechanism of action of exosomes in promoting cancer cell growth and elevating cisplatin resistance is clinically significant.
Protective autophagy is a presurgical mechanism that sequesters, breaks down, and recycles proteins and organelles within lysosomes, preserves mitochondrial integrity, mitigates DNA damage, and reinforces the metabolic fitness and endurance of cancer cells.[9] In advanced NPC, tumor cells may suffer from nutrient deficiency, hypoxia, and other stresses. Autophagy helps tumor cells recycle intracellular substances, provide energy and raw materials, and enhance their survivability. Lu’s study revealed that Wnt5a attenuates the level of DNA damage and promotes the radiation resistance of NPC cells through cellular autophagy mediated by the key factor Beclin1.[10] Central to cell proliferation, metabolism, and autophagic regulation, the dysregulation of the mammalian target of rapamycin (mTOR) pathway has been implicated in the chemo-resistance mechanisms of numerous cancers.[11] Ras homologue enriched in brain (Rheb), a crucial activator of the mTOR complex 1 (mTORC1), significantly influences mTOR signaling output, which in turn, governs autophagic activity.[12] Emerging evidence suggests that exosomes derived from cancer-associated fibroblasts (CAFs) can stimulate multiplication in tumor cells and foster resistance against chemo-therapeutics such as cisplatin.[13] This investigation aimed to explore the intricate process by which CAF-derived exosomes facilitate cisplatin resistance in NPC through modulation of the Rheb/mTOR signaling cascade and stimulation of autophagy. Uncovering such mechanisms not only promises to illuminate novel aspects of cisplatin resistance in NPC but also paves the way for the development of targeted resistance-reversal strategies and furnishes a robust scientific foundation and potential therapeutic targets. This endeavor holds substantial implications for advancing precision medicine in NPC and merges scientific insight with clinical relevance.
MATERIAL AND METHODS Cell cultureThe NPC cell line 5-8F was obtained from Immocell Biotechnology (cat. IM-H132, Shanghai, China) with short tandem repeat (STR) identification correct, and tests for mycoplasma were negative after cell resuscitation. CAFs were isolated and purified from surgical tumor tissues of patients with NPC, and the purified CAFs cells underwent STR identification and mycoplasma detection. The cell isolation procedure was as follows: Tumor and adjacent normal tissue samples were cut into 1 mm3 pieces and incubated in serum-free Dulbecco’s modified eagle medium (DMEM) (cat. 12430054, Invitrogen; Thermo Fisher Scientific, Waltham, MA) with 37.5 mg/mL of type IV collagenase (cat. 17104019, Thermo Fisher Scientific, Waltham, MA) at 37°C for 2 h. Centrifugation at 4°C, 2,000 rpm for 10 min was performed, followed by supernatant removal. The cell pellet was resuspended in phosphate-buffered saline (PBS), filtered through a 400-mesh sterile filter (cat. 352340, Falcon, Corning, New York, USA), and centrifuged again, and the supernatant was discarded. Cells were then plated in DMEM high-glucose medium (10% fetal bovine serum) at 37°C, 5% CO2. After 3 h, the medium was replaced, and nonadherent cells were washed off with PBS before continuing the culture. Fresh medium was changed as needed, and upon reaching 80–90% confluence, the cells were passaged with 0.25% trypsin (cat. C0201, Beyotime, Shanghai, China). This study was approved by the ethical committee (No: LLW-FO-401) and conducted in accordance with the Declaration of Helsinki for experiments involving humans, and all patients signed informed consent.
Methylthiazolyldiphenyl–tetrazolium (MTT)First, 0.5 mL of 5.0 × 106 cells/mL 5-8F cells was cocultured with 0.5 mL of 1 × 105 cells/mL CAFs. Some of the cells were treated with GW4869 (5 μM, cat. S7609, Selleck, Houston, Texas, USA), and the cells were seeded in a 6-well plate. After 24 h, the cells were treated with cisplatin (cat. S1166, Selleck, Houston, Texas, USA) at gradient concentrations for 72 h. Then, the cells were incubated with 100 μL of 0.5 mg/mL (MTT; cat. 298-93-1, Sigma, Darmstadt, Germany) in DMEM medium for 4 h. Next, 150 μL of dimethyl sulfoxide (DMSO) was added to dissolve formazan crystals by shaking. Optical density values were measured at 490 nm using a microplate reader (cat. A51119500C, Thermo Fisher Scientific, Waltham, USA). The survival rates of cells under different cisplatin concentrations were evaluated as mean absorbance of the treatment group/mean absorbance of the control group × 100%, and a dose-effect curve was plotted. The IC50 value was defined as the concentration that causes 50% inhibition of cell proliferation and calculated using nonlinear regression analysis in GraphPad Software 8.00 (GraphPad Software, Inc. San Diego, CA, USA).
5-Ethynyl-2’-deoxyuridine (EdU) stainingThe 5-8F cells treated with different drugs were incubated with 50 μM of EdU reagent (cat. C10310, RiboBio, Guangzhou, China) for 2 h, then incubated with hoechst 33342 solution (C1028, Beyotime, Shanghai, China), and observed by a Nikon Eclipse 80i microscope (cat. 9373261, Nikon, Tokyo, Japan).
Exosome isolation and identificationExosomes were isolated by a differential centrifugation procedure at 4°C: (1) 500 × g for 10 min to remove cells, (2) 2,000 × g for 20 min to remove cellular debris, (3) 10,000 × g for 30 min to remove large vesicles, (4) 110,000 × g for 70 min to remove the supernatant and resuspend in PBS, and (5) 110,000 × g 70 min to purify the exosomes. The exosomes were identified through transmission electron microscopy (TEM; JEOL JEM 1210, JEOL Ltd., Peabody, MA, USA), nanoparticle tracking analysis (NTA), and Western blot (WB) analysis of exosomal markers. 1. TEM: 10 μL of exosomes were fixed with 1% glutaraldehyde (cat. G6257, Sigma, Shanghai, China) and applied to copper grids, then stained with 50 μL of uranyl acetate (cat. SPI 02624, HEAD Biotechnology, Beijing, China) for 5 min, excess liquid was removed, air dried for 5–10 min, and imaged under 80 kV TEM. 2. NTA Particle Size and Concentration Analysis: Using a clean, disposable sample chamber, the outer wall was wiped to prevent contamination adherence. With gentle tilting, the exosome solution was slowly pipetted along the wall to avoid bubbles, covered with a thin lid, and inserted into a Zetasizer Nano ZS 90 particle size analyzer (Malvern Panalytical, Worcestershire, UK). Following standard protocols, the particle size distribution and concentration of the exosomes were analyzed.
Exosome transfection and cellular uptake assayExosomes derived from CAFs were introduced into NPC cells via direct coculture. 5-8F cells were digested and counted, and 105 cells were cocultured with 20 μg of exosomes for 24 h prior to further experimentation. The PKH 26 Red Fluorescent Cell Linker Kit (cat. CGLDIL, Sigma, Darmstadt, Germany) was utilized for lipid bilayer staining. The exosomes were resuspended in 100 μL of Diluent C and incubated with 100 μL of PKH26 dye for 5 min, and the reaction was stopped with 500 μL of serum. Labeled exosomes were washed with PBS and incubated with recipient cells in a 12-well plate for 8 h before imaging. Finally, the intracellular localization of exosomes was observed under a fluorescence microscope (cat. 9373261, Nikon, Tokyo, Japan).
Monodansylcadaverine (MDC) fluorescence assayDifferent treated 5-8F cells in the log phase were incubated with MDC dye (cat. C3018S, Beyotime, Shanghai, China) for 30 min away from light. The wash buffer was diluted with double distilled water, and the cells were gently washed. An appropriate amount of wash buffer was added, and the cells were observed under a fluorescence microscope.
Cell colony formation assayCells were inoculated in culture dishes at a density of 1,000 cells per well. After 7 days of incubation, the cells were fixed, an appropriate amount of crystal violet (cat. C0121, Beyotime, Shanghai, China) was added, and the cells were stained for 15–30 min at room temperature. Then, the staining solution was removed, and the dishes were dried to calculate the number and size of tumor spheres.
Western Blot (WB)After harvesting the cells, they were lysed with RIPA lysis buffer (cat. 89900, Thermo Fisher Scientific, Waltham, USA) for 10 min at 4°C. Proteins were separated using 10–15% polyacrylamide gels and transferred to PVDF membranes (cat. IPFL00010, Millipore, Darmstadt, Germany). Blots were blocked with 5% bovine serum albumin (BSA) (cat. A8020, Solarbio, Beijing, China) for 1 h at room temperature and incubated with primary antibody overnight at 4°C. GAPDH was used as a control. Then, the blot was incubated with secondary antibody horseradish peroxidase-conjugated goat antirabbit (1:5,000; cat. ab205718, Abcam, Cambridge, UK) or goat antimouse (1:5,000; cat. ab205719, Abcam, Cambridge, UK), and the signal was observed using an Odyssey infrared imaging system. The following antibodies were used: tumor susceptibility gene 101 (1:5,000; ab125011, Abcam, Cambridge, UK), cluster of differentiation (CD)63 (1:2,000; ab68418, Abcam, Cambridge, UK), CD81 (1:2,000; ab109201, Abcam, Cambridge, UK), light chain (LC)3B (1:1,000; 18725-1-AP, Proteintech, Wuhan, China), GAPDH (1:5,000; GB11113-100, Servicebio, Wuhan, China), Rheb (1:2,000; T58376, Abmart, Shanghai, China), Beclin-1 (1:1,000; AB207612, Abcam, Cambridge, UK), mTOR (1:1,000; 28273-1-AP, Proteintech, Wuhan, China), and UNC51-like kinase (ULK1) (1:1,000; 29005-1-AP, Proteintech, Wuhan, China).
Quantitative real-time polymerase chain reaction (qRTPCR)Total RNA was extracted by Trizol (cat. 15596026CN, Invitrogen, Shanghai, China), evaluated for RNA purity, and reverse transcribed to cDNA. An SYBR@ GreenERTMqPCR reaction system (cat. 4309155, Thermo Fisher Scientific, Waltham, USA) was constructed, and GAPDH was used as the internal reference gene. The relative expression of the genes to be evaluated was calculated by 2-∆∆CT. The primer sequences were as follows: Rheb F 5’-GAGTCCACAAATTGGCCTTC-3’, R 5’-CAGTCCAAGTCCCGGAAGAT-3’; mTOR F 5’-ATG CTTGGAACCGGACCTG-3’, R 5’-TCTTGACTCATCTCT CGGAGTT-3’; LC3 F 5’-GAGTGGAAGATGTCCGGCTC-3’, R5’-CCAGGAGGAAGGCTTGG-3’; ULK1 F 5’-CCACCCAG TTCCAAACACCTR-3’, 5’-CCAACTTGAGGAGATGGCG T-3’; BECN1 F5’-GAGGGATGGAAGGGTCTA-3’, R 5’-GCC TGGGCTGTGGTAAGT-3’; GAPDH F 5’-GTGGCCGAG GACTTTGATTG-3’, R 5’-CCTGTAACAACGCATCTCATA TT-3.’
ImmunofluorescenceCells were washed twice with PBS, fixed with 4% paraformaldehyde (cat. 441244, Sigma, Shanghai, China) for 15 min, and permeabilized with 0.5% Triton X-100 (cat. T8787, Sigma, Shanghai, China) for 30 min at room temperature. After being closed with 3% BSA for 40 min, the cells were incubated with mTOR antibody (1:500) overnight at 4°C. The cells were then incubated with the fluorescein isothiocyanate -labeled secondary antibody (1:200, Abclonal Tech, Wuhan, China) for 1 h. The localization and expression intensity of mTOR in the cells were photographed under a fluorescence microscope using Nikon NIS-Elements software (Nikon, Tokyo, Japan).
Tumor transplantation in miceA total of 18 male AthymicC57 nude mice of 4–6 weeks of age were purchased from Shanghai Laboratory Animal Center, China. Preparations of 3 × 105 5-8F cells or 5-8F cells cocultured with CAFs cells were suspended in 200 μL of serum-free DMEM and injected subcutaneously into each mouse. For the exosome group, mice in the 5-8F cells group were injected into the tail vein with 20 μg of exosomal protein. After 10 days, intraperitoneal injections of 10 mg/kg of cisplatin-diamminedichloroplatinum (DDP) or PBS were administered every 4 days, and the mice were sacrificed after 150 mg/kg pentobarbital sodium anesthesia on day 30 for further analysis. All mice were kept in pathogen-free cages. The animal research was in accord with the animal welfare ethical review and examined and approved by the animal ethics committee.
Statistical analysisAll data were expressed as mean ± standard error. Statistical analysis was performed using GraphPad Prism 8.00 (GraphPad Software, Inc. San Diego, CA, USA). Statistical significance was analyzed using Student’s t-test or one-way analysis of variance, followed by a Tukey’s test for post hoc testing. P < 0.05 was considered statistically significant.
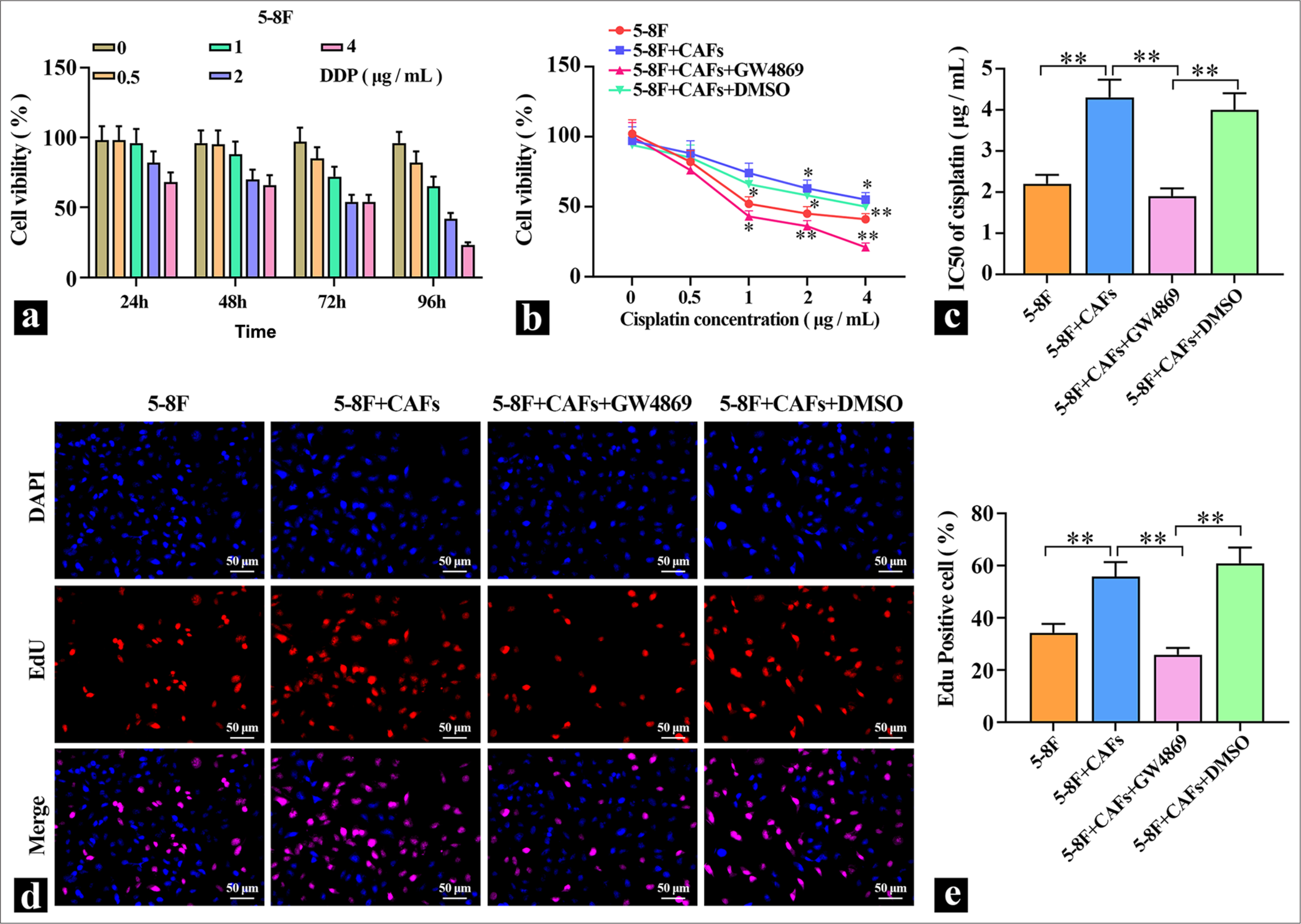
RESULTS Coculture of CAFS cells with 5-8F promoted the proliferation and increased DDP resistance of 5-8F cellsTo detect the effects of DDP on NPC cells, different concentrations of DDP were applied to 5-8F cells for varying durations, and cell viability was measured. The results in Figure 1a show that DDP reduced the viability of 5-8F cells in a dose- and time-dependent manner, and DDP inhibited the proliferation capacity of 5-8F cells. Treatment with 2 μg/mL of DDP for 72 h resulted in a significantly decreased cell viability in 5-8F cells, which were used in subsequent experiments. Then, CAFs were directly cocultured with 5-8F cells, and the coculture system was treated with 5 μM of the exosome inhibitor GW4869. The results in Figure 1b and c show that compared with the 5-8F cell group, the coculture of CAFs with 5-8F cells led to a significant increase in cell viability and a marked elevation in the IC50 value for DDP, which indicated a clear enhancement of 5-8F cell resistance to DDP. After adding the exosome inhibitor, cell viability decreased significantly, which suggested that the promotion of 5-8F cell proliferation by CAFs might be mediated through the release of exosomes. To validate the role of exosomes further, EDU staining was performed to detect cell proliferation. The staining results depicted in Figure 1d and e show that coculturing CAFs with 5-8F cells significantly enhanced their proliferation ability. When the secretion of exosomes from CAFs was inhibited, the proliferation ability of 5-8F cells in coculture was markedly reduced, which indicated that exosomes derived from CAFs served a key role in promoting cell proliferation and elevating cellular resistance to DDP.
Export to PPT
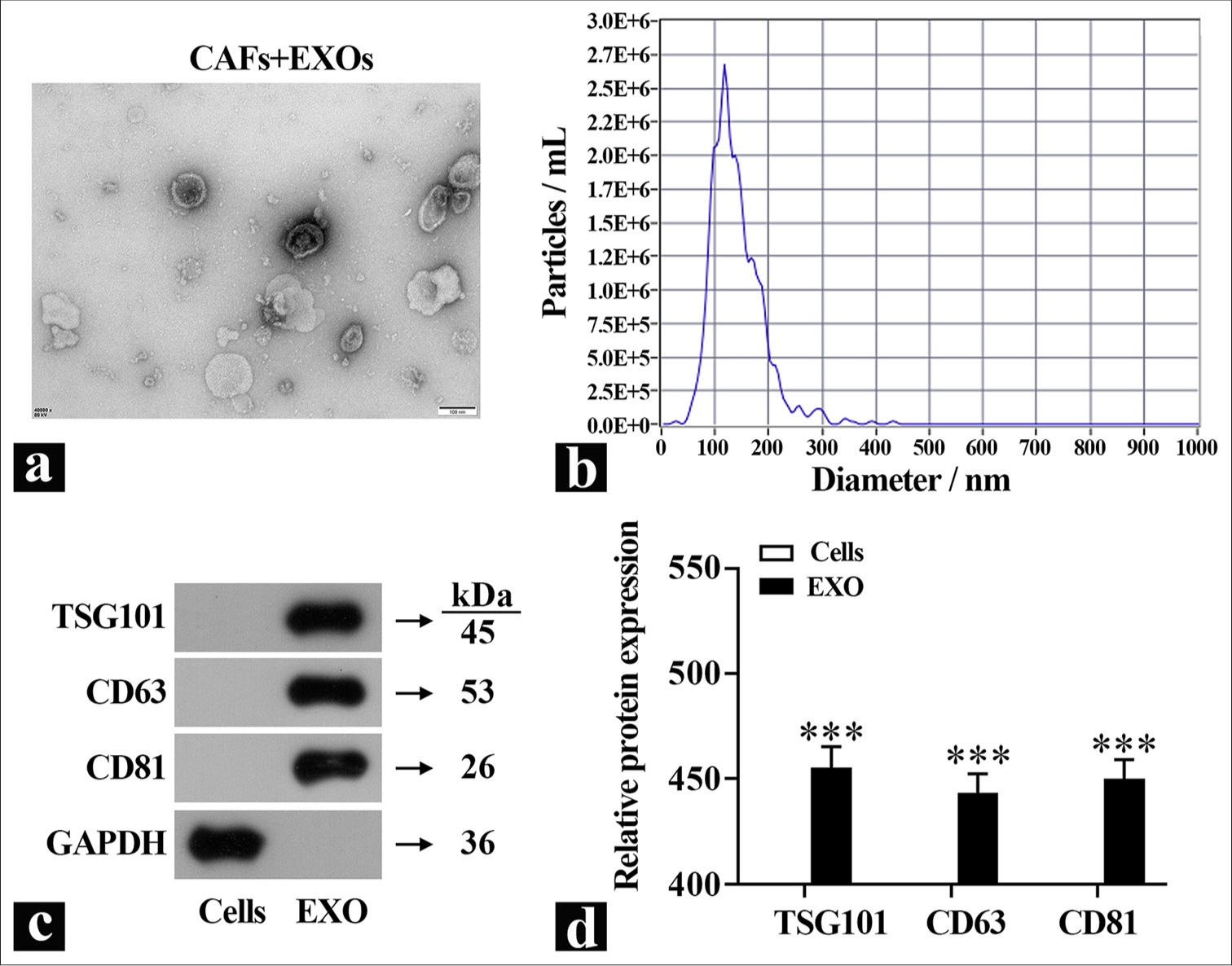
Isolation and characterization of primary CAF-derived exosomesTo investigate the functions of exosomes further, they were isolated from primary CAFs using differential centrifugation, and the exosome morphology was observed using TEM [Figure 2a]. Particle size analysis revealed that the diameter of exosomes derived from CAFs was around 100 nm [Figure 2b]. Quantification of exosomes marker proteins in the isolated exosomes showed minimal amounts of these markers in the cell lysate group [Figure 2c and d], which indicated the successful isolation of the exosomes.
Export to PPT
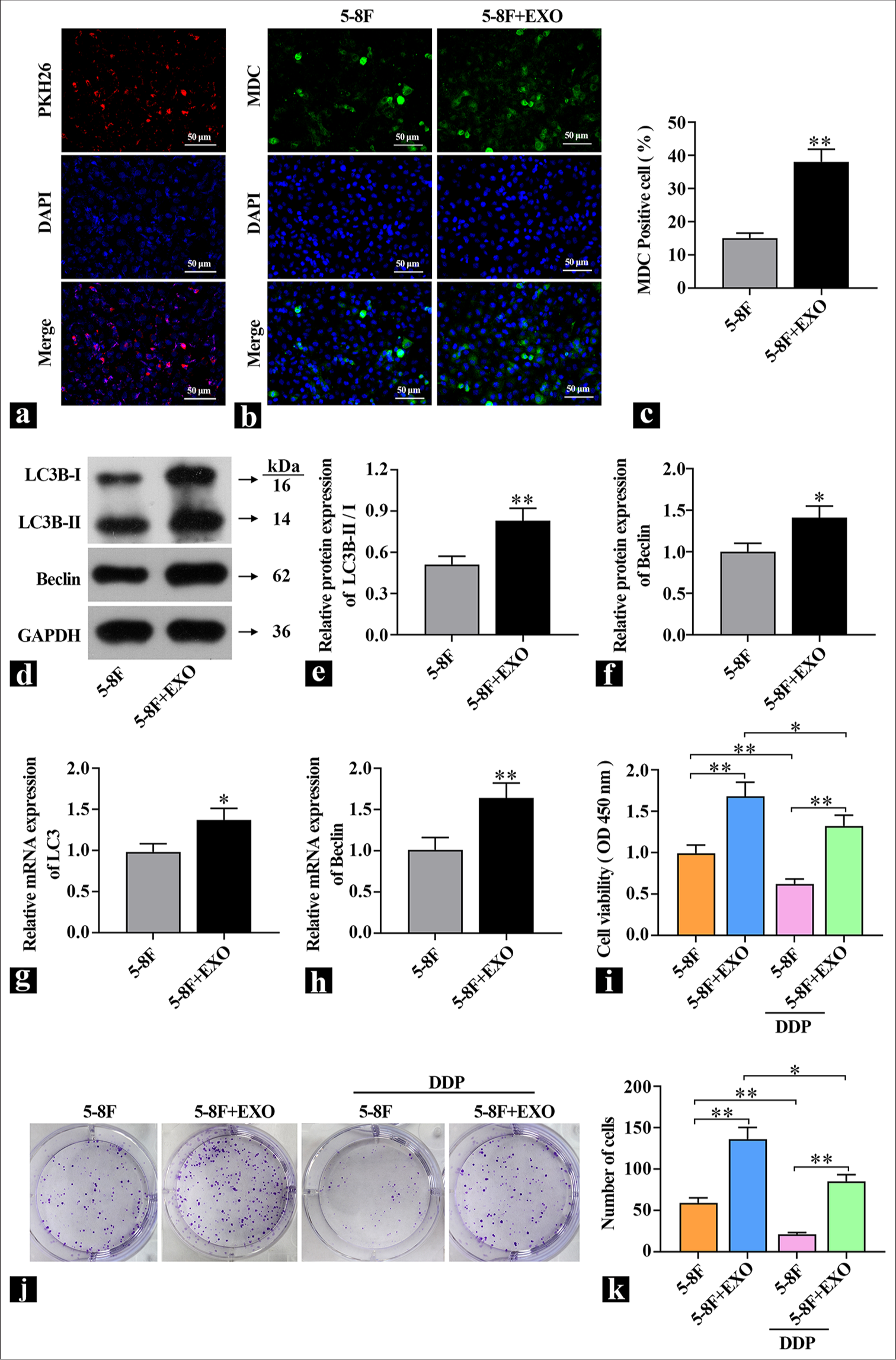
CAF-derived exosomes enhance autophagy and promote DDP resistance in 5-8F cellsThe isolated exosomes derived from CAFs were introduced into 5-8F cells. An exosome labeling (KH26) and tracing experiment was conducted to assess the transfection efficiency. The results in Figure 3a show that the red fluorescence of PKH26 overlapped with nuclear fluorescence and the delivery of exosomes into 5-8F cells was successful. To investigate whether exosomes affect autophagy, changes in the expression of autophagy marker proteins LC3B and Beclin, as well as MDC staining, were used to detect autophagy flux. Compared with the control group, the MDC fluorescence intensity was elevated [Figure 3b and c], and the expression of autophagy marker proteins LC3B-II/I and Beclin was also significantly increased in the exosome group [Figure 3d-h], which indicated the administration of exosomes significantly increased the level of autophagy in 5-8F cells. MTT and colony formation assays were used to detect the effect of exosomes on DDP resistance. Figure 3i shows that compared with the control group, the cell viability was significantly increased in the exosome group; after the addition of DDP, cell viability significantly decreased in the control group, but the difference was reduced compared with exosomes without DDP. The colony formation assay results in Figure 3j and k show that after DDP treatment, compared with the 5-8F cell group, the introduction of exosomes into 5-8F cells led to an increase in the number of colonies formed, which indicated that 5-8F cells had a protective response against DDP damage. These findings suggested that coculturing exosomes with NPC cells promoted autophagy levels and enhanced resistance to DDP.
Export to PPT
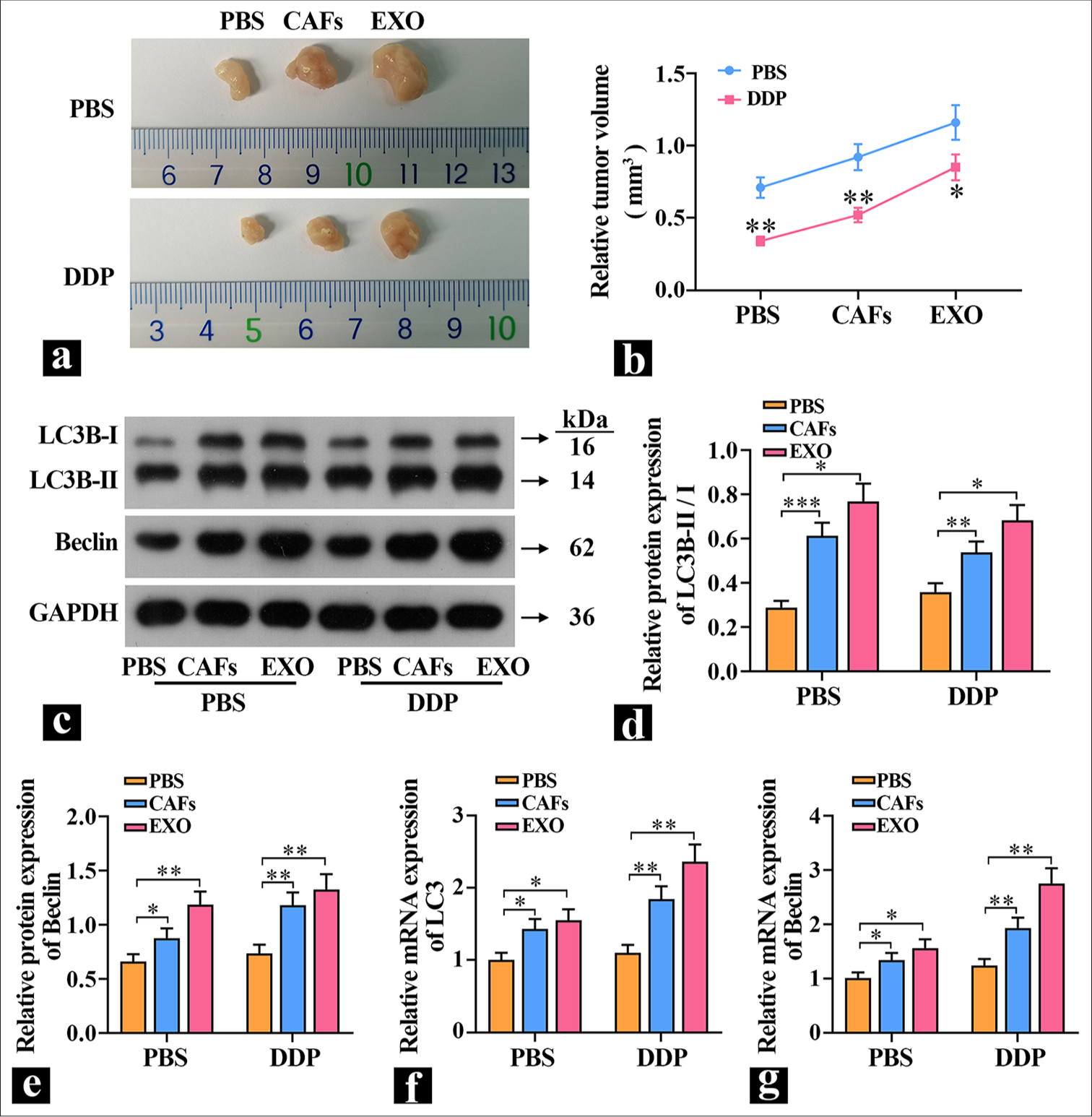
CAF-derived exosomes increase autophagy and promote tumor growth in mice5-8F cells were either cultured alone or cocultured with CAFs and then implanted subcutaneously into mice. The exosome group received exosome proteins, and the effects of exosomes on tumor growth were observed by measuring the tumor volume in each group of mice. The results in Figure 4a and b show that CAFs and exosomes significantly promoted tumor growth. Treatment with DDP caused a significant reduction in tumor volume in the PBS group. Compared with the PBS group, the tumor volume in the exosome group was significantly larger, which indicated that exosome proteins could significantly promote tumor growth in vivo and decrease tumor cell sensitivity to DDP. Subsequently, the expression levels of autophagy proteins were detected. The results in [Figure 4c-g] are similar to those in vitro: Compared with the PBS group, the expression of LC3B-II/I was significantly higher in the exosome group, and the expression level of Beclin was extremely significantly increased, which suggested that exosomes can elevate autophagy levels and promote tumor growth.
Export to PPT
Exosomes promote protective autophagy and increase DDP resistance by regulating Rheb/mTORTo explore the specific way that exosomes regulate autophagy, NPC cells were pretreated with 1 μM of autophagy promoter rapamycin and then treated with exosome and DDP. The protein and mRNA levels of Rheb, mTORC1, and ULK1 were detected by WB or qRT-PCR [Figure 5a-g]. Compared with the control group, the mTOR inhibitor rapamycin decreased the expression levels of Rheb and mTORC1 and increased the expression levels of ULK1. The cells with exosomes added had the same effect as rapamycin. After rapamycin pretreatment, the expression levels of Rheb and mTORC1 were further increased, and the expression levels of ULK1 were further decreased. These results indicate that exosomal proteins acted on the upstream of mTOR and inhibit mTOR expression. Subsequently, the effect of DDP on DDP sensitivity was observed. The cell viability results show that DDP decreased the cell viability of the control group, the cell viability of the rapamycin group and the exosome group increased significantly, and the cell viability of the coacting group increased significantly [Figure 5h]. The localization and expression intensity of mTOR in cells were observed by immunofluorescence using mTOR antibodies. Compared with the 5-8F group, the fluorescence intensity of the rapamycin group and the exosome group was significantly reduced, which suggested that exosomes inhibited mTOR activity [Figure 5i and j]. In the cell colony experiment, DDP inhibited cell proliferation in the control group, whereas the rapamycin group and the exosome group increased cell proliferation [Figure 5k and l], which indicated that one of the important ways for exosome to increase the resistance of cells to DDP is to improve cell protective autophagy.
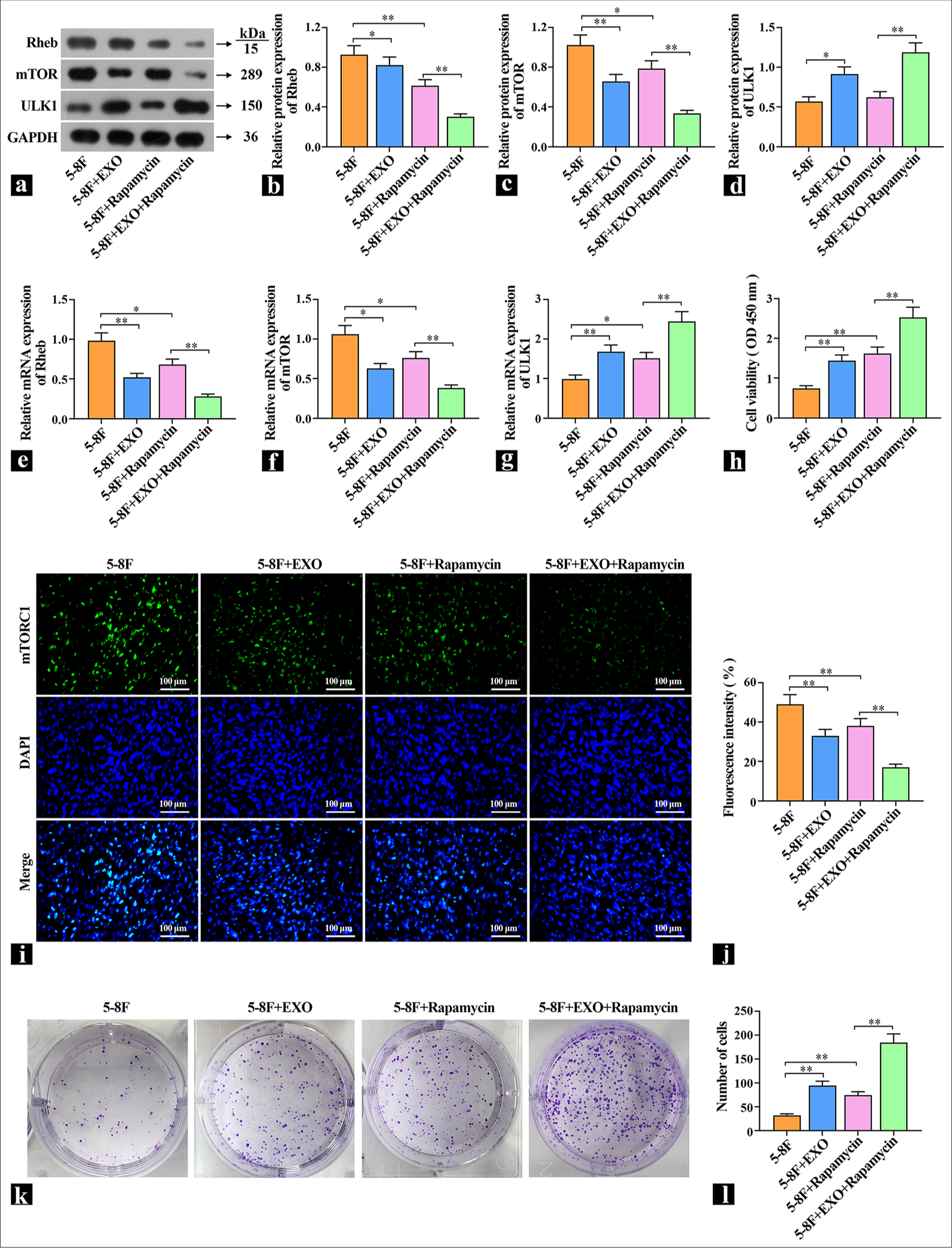
Export to PPT
DISCUSSIONNPC typically goes undetected until advanced stages due to the absence of clear early indicators, which results in an poor prognosis for 60% of diagnosed patients.[14] Clinically, concurrent platinum-based chemo-radio therapy is the standard treatment for locally advanced NPC.[15] Disease progression correlates with a decline in cancer cells’ responsiveness to cisplatin and exacerbates patients’ prognoses.[16] The TME, an intricate ecosystem consisting of resident malignant cells, infiltrating immune cells, metabolic byproducts, and extracellular matrices, actively contributes to the development of drug resistance in NPC cells. Consequently, exploring the mechanisms of resistance tied to the TME in late-stage NPC can pave the way for identifying novel targets capable of reverting cisplatin resistance.
CAFs, ubiquitous in solid tumors, stand out within the TME’s diverse cellular milieu due to their capacity to govern tumor cell multiplication, invasion, and foster therapeutic resilience and immune escape through multifaceted mechanisms.[17] High CAF densities correlate inversely with patient survival and implicate their carcinogenic role.[18] Huang’s findings illuminated CAFs’ capability to enhance radiotherapy-treated NPC cell survival.[19] Exosomes serve as conduits ferrying bioactive molecules between cells and thereby influence cellular dynamics within the tumor milieu. They partake in processes pivotal to tumor progression such as cell proliferation, angiogenesis, metastasis, immunosuppression,[20] and chemo-resistance. Specifically, exosomes shed by nasopharyngeal CAFs mediate drug resistance in recipient carcinoma cells via constructing a tumor-supportive micro environment [21] and impeding immune surveillance.[22] Exosomes facilitate the transfer of cisplatin resistance, and exosomes from doxorubicin-resistant endothelial cells amplify invasiveness, metastasis, epithelial–mesenchymal transition, and chemo-resistance in NPC.[23] This study revealed that when CAFs and NPC cells (5-8F) were cocultivated, a marked increase in carcinoma cell proliferation and decrease in cisplatin resistance ensued. However, upon application of exosome inhibitors during coculture, the promotional effect was notably abrogated, which highlighted the pivotal role of CAF-derived exosomes in these processes.
Growing evidence suggests that suppressing autophagy could emerge as a viable therapeutic strategy for advanced cancers;[24] it initially functions as a tumor suppressor by mitigating tissue injury and inflammation, and preserving genomic stability through the degradation of carcinogenic stimuli and management of oxidative stress. Conversely, in the later stages of tumor, autophagy facilitates nutrient supply to cancer cells, fosters their immune evasion, and thereby aids tumor progression.[25] NPC research has demonstrated elevated autophagy levels in these cells, which not only aids in their survival environmental stress, but also bolsters proliferation, invasiveness,[14] and confers radio-resistance, Thus, targeting protective autophagy in cancer cells emerges as a pivotal strategy in cancer control. In our experimental context, exosomes secreted by CAF-augmented protective cell proliferation via stimulation of autophagy concurrently heighten the cells’ resilience against cisplatin treatment.
mTOR, acting as a central regulator of autophagy initiation, primarily operates through mTORC1. By directly phosphorylating and thereby inhibiting the Atg1 (ULK1) complex, mTORC1 negatively controls autophagic activity. mTOR suppression has been implicated in chemotherapy persistence and attenuation of chemotherapeutic susceptibility.[26] In the present investigation, the treatment of 5-8F cells with the mTOR inhibitor rapamycin resulted in an elevated expression of Rheb and mTORC1, along with a decrease in ULK1 levels. Following exposure to exosomes, Rheb and mTORC1 expressions were augmented even further, whereas ULK1 levels experienced additional reduction, which suggested exosome induction of protective autophagy via mTOR down-regulation. Further exploration confirmed that rapamycin treatment and exosomal exposure suppressed mTORC1 expression in cells and reinforced the proposed mechanism: CAF-derived exosomes enhance autophagy and diminish cisplatin sensitivity through mTOR inhibition.
Cisplatin may exert dual roles in its treatment of cancer cells. In many cases, cisplatin therapy promotes autophagy in cisplatin-sensitive and cisplatin-resistant cells.[27] Therefore, enhancing autophagy is considered a strategy for improving cisplatin chemo-sensitivity and treating cancer. However, the functional activity of cisplatin-induced autophagy is associated with different genetic phenotypes, tumor types, and the TME.[28] While cisplatin combats cancer, it can trigger protective autophagy in cells, such as by inhibiting the mTOR signaling pathway, thus relieving mTOR’s suppression of autophagy and initiating autophagy. Future research should delve deeper into the balance between cisplatin and the regulation of autophagy levels to enable cisplatin to achieve more effective therapeutic outcomes in clinical treatments.
SUMMARYThis study explored the mechanism by which CAF-derived exosomes promote the protective cell autophagy of NPC cells through Rheb/mTOR, thus leading to increased cisplatin resistance and providing a theoretical basis for further research on drug resistance of NPC and directions for the development of new therapeutic strategies.
Comments (0)