Cervical cancer persists as a considerable global health issue, standing as a primary contributor to morbidity and mortality associated with cancer in women.[1-3] While progress has been made in preventive measures, diagnostics, and therapeutic approaches, it is essential to understand the molecular mechanisms driving the progression of cervical cancer. This knowledge is crucial for the development of targeted therapies and enhancing patient outcomes.[4]
Mitochondria, essential cellular organelles, are integral in energy generation, maintaining redox balance, and influencing decisions related to cell fate.[5,6] Dysfunction in mitochondrial processes has been implicated in various cancers, and understanding the specific contributions of mitochondrial proteins to cancer progression is an area of active research.[7,8]
One such mitochondrial protein of interest is Isoleucyl-tRNA Synthetase 2 (IARS2), a member of the aminoacyl-tRNA synthetase family.[9] While traditionally recognized for its role in protein synthesis, emerging evidence suggests that IARS2 may have additional functions within the mitochondria, impacting cellular processes beyond its canonical role.[10]
In the context of cervical cancer, the influence of IARS2 on mitochondrial function, oxidative stress, and cell proliferation remains poorly characterized. Investigating the interplay between IARS2 and these cellular processes could provide valuable insights into the molecular mechanisms driving cervical cancer pathogenesis.
The objective of this research is to investigate how the mitochondrial protein IARS2 affects mitochondrial function, oxidative stress levels, and cellular proliferation in cervical cancer cells. By conducting a thorough analysis of these interrelated facets, our goal is to reveal potential molecular targets for therapeutic interventions and contribute to a more profound comprehension of the intricate biology driving the progression of cervical cancer.
MATERIAL AND METHODS Source of clinical specimensCollect surgical specimens from cervical cancer patients in the Obstetrics and Gynecology Department of the Zibo Central Hospital, from the same location as the specimens collected by the Pathology Department, confirmed by pathological diagnosis. Immunohistochemical specimens are immediately placed in 10% formalin fixation. The adjacent tissue of cervical cancer is the adjacent tissue of focal tumor or cervical tissue from patients undergoing hysterectomy due to benign lesions. There are a total of 30 pairs of matched cervical cancer and adjacent tissue specimens in this study. Informed consent forms have been signed by all cervical cancer patients included in this study.
ImmunohistochemistryFirst, tissue specimens are fixed and subjected to dehydration, embedding, and sectioning. Subsequently, the sections undergo antigen retrieval to restore and expose protein antigens, as well as to block non-specific binding. Next, the samples are incubated with a specific primary antibody (IARS2) (dilution: 1:100, ab242116, Abcam, Cambridge, UK), followed by washing to remove unbound primary antibody. Then, the samples are incubated with a secondary antibody labeled with specific detection markers to achieve specific binding. Optionally, signal amplification reagents can be used to enhance detection signal intensity. Staining substrate is applied to the sections to generate visible staining reactions where specific binding of the secondary antibody (dilution: 1:100, ab288151, Abcam, Cambridge, UK) occurs in the tissue. Following staining, the sections are dehydrated and washed, and finally covered with mounting medium to protect and facilitate microscopic observation. Expression of the target protein can be assessed by observing and analyzing the stained sections under a light microscope.
Cell cultureWe purchased the normal human cervical epithelial cells (HCECs) (CP-H059) and cervical cancer cell lines (HeLa cells) (CL-0101) from Procell (Wuhan, China). Following procurement, the cells were cultured in Roswell Park Memorial Institute (RPMI) 1640 complete medium (iCell-0002, iCELL, Shanghai, China) (with 10% fetal bovine serum [FBS]) and maintained at 37°C with 5% Carbon dioxide (CO2). Passaging occurred every 3–4 days utilizing a 0.1% trypsin solution. On entering the logarithmic growth phase, the cells were subjected to trypsin digestion, collected through centrifugation, quantified, and suspended in serum-free RPMI 1640 medium at a concentration of 1 × 107 tumor cells/ml for subsequent procedures. All cell lines employed in this investigation underwent short tandem repeat identification and mycoplasma testing.
Short hairpin RNA (ShRNA) and plasmid transfectionInitially, Hela cells were cultivated in a 6-well plate and permitted to incubate for a period of 24 h. Subsequently, the cells were transfected using Lipofectamine 2000 (Thermo Fisher Scientific, Beijing, China) with ShRNA (negative control), ShRNA (IARS2), pcMV-FLAG plasmid, and pcMV-FLAG-IARS2 plasmid (pcMV-FLAG-IARS2) (Sangon, Shanghai, China). Following transfection, the cell culture medium was refreshed every 6 h, repeating the process a total of 8 times. Gene expression analysis in Hela cells, encompassing messenger Ribonucleic Acid (mRNA) and protein levels, was conducted through quantitative reverse transcription polymerase chain reaction (qRT-PCR) and western blot. Detailed information on the interference fragment sequences utilized in these experiments is provided in Table 1.
Table 1: The gene interference fragment sequences involved in this chapter.
ShRNA/Plasmid names Insert sequence(5’-3’) ShRNA-NC-sense GCACTACCAGAGCTAACTCAGATAGTACT ShRNA-NC-antisense AGTACTATCTGAGTTAGCTCTGGTAGTGC ShRNA-IARS2-sense GCTGCATCCTTACCTACAA ShRNA-IARS2-antisense TTGTAGGTAAGGATGCAGC pcMV-FLAG-IARS2-sense GAATTCATGCTCTTCCCCCGGCCGGCTGGAAGAGCCCGGCCAGCGCGGCIn summary, the extraction of total cellular/tissue RNA was carried out using TRIzol reagent. Subsequently, RNA (1.0 μg per reaction) underwent reverse transcription employing the Two-Step RT-PCR Kit (Takara Bio, Japan). Real-time qRT-PCR was conducted utilizing the 7900 polymerase chain reaction (PCR) system (Applied Biosystems, Shanghai, China) in conjunction with SYBR GREEN PCR Master Mix (Thermo Fisher Scientific). Consistently, glyceralde-hyde-3-phosphate dehydrogenase (GAPDH) mRNA expression served as the internal control for examination. The calculation of relative mRNA expression levels was performed using the 2-ΔΔCt method. The specific primers designed for this study are presented in Table 2.
Table 2: The primer sequences utilized in the current investigation.
Primer name Primer sequences(5’-3’) IARS2-F TGGACCTCCTTATGCAAACGG IARS2-R GGCAACCCATGACAATCCCA GAPDH-F GAAGGTGAAGGTCGGAGTC GAPDH-R GAAGATGGTGATGGGATTTC Western blot techniqueProteins were extracted using radioimmunoprecipitation assay lysis buffer, followed by separation through sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and subsequent transfer onto a polyvinylidene difluoride membrane. The membrane was then blocked with 5% bovine serum albumin. Primary antibodies, specifically Glyceraldehyde 3-Phosphate Dehydrogenase (GAPDH) (dilution: 1:2000, ab9485, Abcam, Cambridge, UK), IARS2 (dilution: 1:1000, ab242116, Abcam, Cambridge, UK), Cleaved caspase-3 (dilution: 1:1000, ab2302, Abcam, Cambridge, UK), Bax (dilution: 1:1000, ab32503, Abcam, Cambridge, UK), and Bcl-2 (dilution: 1:1000, ab182858, Abcam, Cambridge, UK), were left to incubate overnight at 4°C. Subsequently, the membrane underwent exposure to Horseradish peroxidaseconjugated secondary antibodies (dilution 1:2000; cat no. ZB-2305 and ZB-2301, ZSGB-BIO) at room temperature for 1 h. Following thorough washing, protein bands were visualized using an electrochemiluminescence system (ChemiDoc XRS+, Bio-Rad ChemiDoc, California, USA). Grayscale analysis was finally conducted using ImageJ software (version 1.5f, NIH, Maryland, USA).
Cell counting kit-8 (CCK-8) viability and Trypan Blue cell death assaysInitially, cervical cancer cells treated with the IARS2 gene were seeded into individual wells of a 96-well plate, with a cell density of 4000 cells per well. Subsequently, the cells were maintained in a 37°C, 5% CO2 incubator for an additional 72 h to allow for proper incubation and cell proliferation. Following this incubation period, a CCK-8 mixture (CA1210, Solaibio, Beijing, China) at a volume of 10 μL per well was added to the cells. After a subsequent 2-h incubation, the absorbance of the CCK-8 solution was measured at 450 nm using a microplate reader (Synergy H1, BioTek Instruments, Winooski, Vermont State, USA). To assess cell death, cervical cancer cells were stained with Trypan Blue (C0040, Solarbio, Beijing, China). The “dead” cells, which took up Trypan Blue, were then quantified using an automated cell counter. This comprehensive approach allows for a thorough evaluation of the impact of IARS2 gene treatment on cell viability and death in cervical cancer cells.
Enzyme-linked immunosorbent assay (ELISA)Acquired the Mito-Complex I Activity Assay Kit (Ab109903, Abcam, Cambridge), adenosine triphosphate (ATP) assay kit (BC0300, Solarbio, Beijing, China), and reactive oxygen species (ROS) Assay Kit (D6470, Solarbio, Beijing, China) for the experimental procedures. Prepared cell extracts and executed the specific experiments meticulously following the guidelines provided by the manufacturer. Conducted precise activity measurements utilizing an ELISA reader (Synergy H1, BioTek Instruments, Winooski, Vermont State, USA) to ensure accuracy and reliability in the obtained data.
Measurement of optical density (OD) values for JC-1 and Reactive Oxygen Species (ROS)Prepare your cell samples according to the experimental setup. Transfer the cell suspension containing JC-1 staining (JC-1 (M8650, Solarbio, Beijing, China) to a microplate, and measure the OD values using a microplate reader at 490 nm (excitation) and 530 nm (emission) for the monomeric form, and at 570 nm (excitation) and 595 nm (emission) for the J-aggregate form. Transfer the cell suspension containing 2’,7’-dichlorodihydrofluorescein diacetate (DCFH-DA) staining (D6470, Solarbio, Beijing, China) to a microplate (Synergy H1, BioTek Instruments, Winooski, Vermont State, USA), and typically measure OD values at 485 nm (excitation) and 535 nm (emission). Analyze the obtained OD values to quantify the relative levels of ROS in each sample.
Transwell assayThe cervical cancer cells, genetically modified with the IARS2 gene, were resuspended at a density of 1.5 × 104 cells per well in serum-free culture medium. Subsequently, these cells were placed on the upper surface of the Transwell chamber. The lower surface of the chamber was filled with complete culture medium containing 12% Fetal Bovine Serum (FBS), facilitating cell migration for a duration of 24 h. Following the migration period, the cells located on the upper surface of the chamber were meticulously wiped away using a cotton swab. The migrated cells on the lower surface were subjected to thorough washing, fixing, and staining procedures. The migrated cells on the lower surface were then observed and documented using a microscope (CKX53, Olympus, Tokyo, Japan). This approach enables a comprehensive assessment of the impact of the IARS2 gene on cervical cancer cell migration.
Fluorescence stainingCells were placed in 6-well plates and cultured normally for 72 h. Then, cells were fixed with 4% paraformaldehyde, washed with phosphate buffer saline (PBS), and permeabilized with 0.5% Triton X-100. Following this, cells were treated with 5-ethynyl-2’-deoxyuridine (EdU; C0071S, Beyotime, Beijing, China) and 2-(4-Amidinophenyl)-6-indolecarbamidine dihydrochloride (DAPI; C1002, Beyotime, Beijing, China). Subsequently, cells were observed under a fluorescence microscope (ZEISS Axio Scan.Z1, Zeiss, Oberkochen, Germany) and cells were counted
Statistical analysis of dataStatistical analyses were performed using GraphPad Prism software version 8.0 (GraphPad Inc., San Diego, California, USA), accessible at https://www.graphpad-prism.cn/. A t-test was employed for comparing two groups, and multiple group comparisons were assessed using one-way analysis of variance, followed by post hoc Tukey tests. The data were presented as mean ± standard deviation, and statistical significance was considered at P-value below 0.05 (P < 0.05).
RESULTS The expression of IARS2 is upregulated in cervical cancerIn the initial phase, we evaluated the expression profile of IARS2 in cervical cancer cells utilizing qRT-PCR and Western blot techniques. The findings depicted in Figure 1a-c reveal a substantial upregulation in both mRNA and protein levels of IARS2 in cervical cancer cells (Hela cells) compared to normal cervical epithelial cells (HCECs) (P < 0.001, and P < 0.01). By immunohistochemical staining, the expression level of IARS2 in tumor tissue is significantly higher than that in adjacent non-cancerous tissue (P < 0.01) [Figure 1d and e]. The Hematoxylin and Eosin (HE) staining results [Figure 1f] show that in the para-cancerous tissue, cell morphology is relatively regular, with consistent cell size, relatively uniform nucleocytoplasmic ratio, evenly distributed chromatin, and relatively rare nuclear division. In tumor tissue, there is a significant enhancement in cellular pleomorphism, with uneven cell size, inconsistent nucleocytoplasmic ratio, uneven distribution of chromatin, and a marked increase in nuclear division. In para-cancerous tissue, glandular structures are relatively intact, with a lower degree of interstitial fibrosis and moderate vascular density. In tumor tissue, glandular structures are disrupted, exhibiting irregular morphology, significantly increased interstitial fibrosis, and increased vascular density. In para-cancerous tissue, the inflammatory response is mild, with minimal infiltration of inflammatory cells. In tumor tissue, the inflammatory response is significantly increased, with abundant infiltration of inflammatory cells, and an active inflammatory response.

Export to PPT
The loss of IARS2 impairs the mitochondrial function of cervical cancer cellsSubsequently, we delved into the impact of IARS2 knockdown on mitochondrial function in cervical cancer cells. Initially, Sh-IARS2 lentivirus was transduced into Hela cells to establish a model featuring the knockdown of IARS2. The outcomes depicted in Figure 2a-c demonstrate a marked reduction in both gene and protein expression levels of IARS2 in the ShIARS2 group compared to the control group and Sh-Negative control (NC) group (P < 0.01). Of notable significance, IARS2 shRNA induced the transition of JC-1 from red fluorescent aggregates to green fluorescent monomers (P < 0.05) as illustrated in Figure 2d and e, indicating that the IARS2 shRNA induced a decline in mitochondrial membrane potential and thus, the impairment in mitochondrial function.
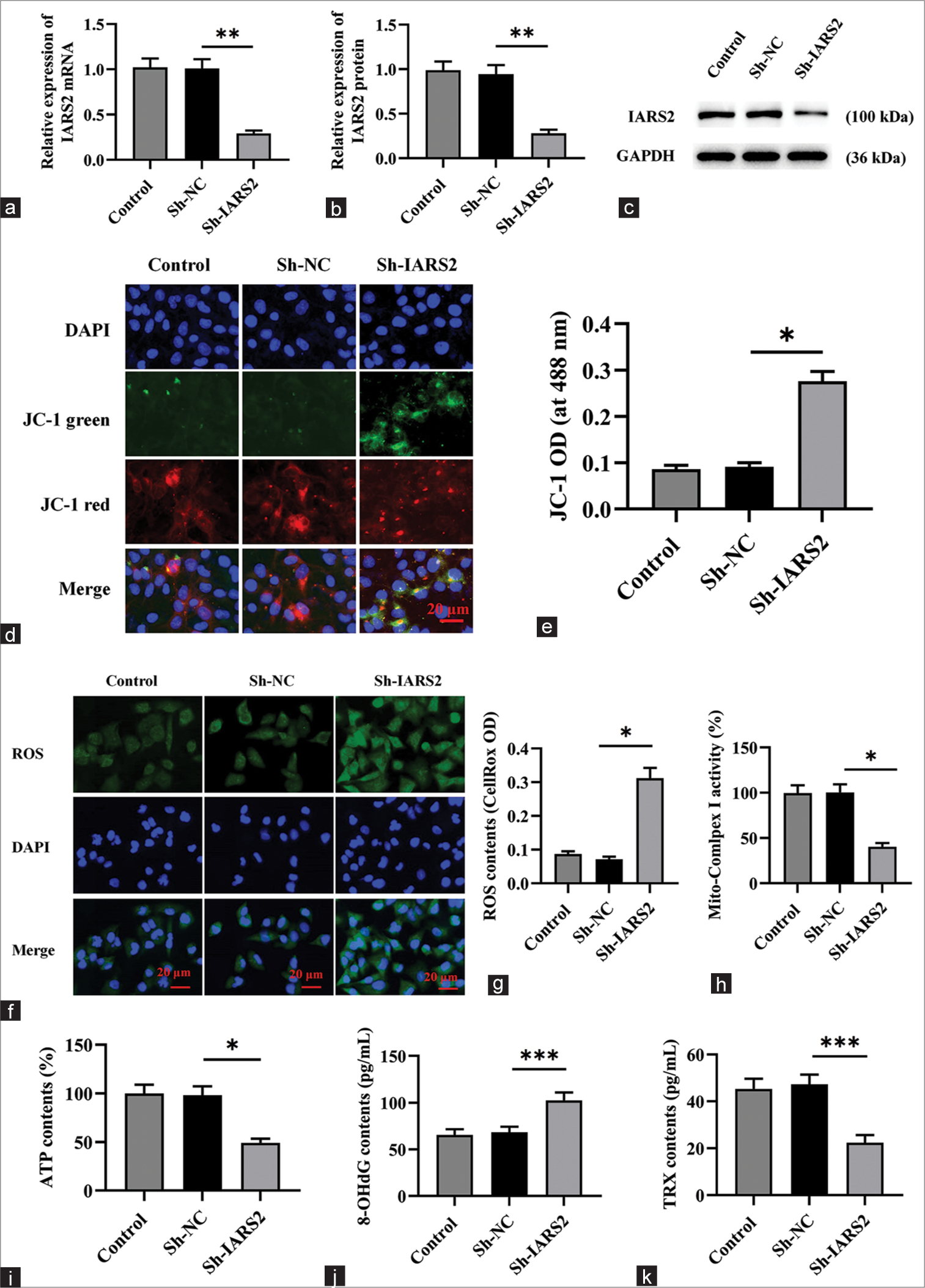
Export to PPT
The elevated green fluorescence intensity observed in Figure 2f and g for the Sh-IARS2 group signifies a substantial increase in ROS production and oxidative stress in Hela cells (P < 0.05). Moreover, IARS2 silencing resulted in a noteworthy decrease in the activity of mitochondrial complex I and a reduction in ATP content within Hela cells (P < 0.05) [Figure 2h and i]. In Figure 2j-k, compared to the Sh-NC group, the levels of 8-OHdG were significantly elevated and the levels of TRX were significantly decreased in the Sh-IARS2 group (P < 0.001).
The knockdown of IARS2 inhibited the survival, proliferation, and migration of cervical cancer cellsSubsequent to the investigation of IARS2 knockdown effects, we proceeded to assess whether this alteration influenced the behavior of cervical cancer cells. The CCK-8 assay was employed to measure cell viability in Hela cells, revealing a substantial reduction in cell viability on IARS2 silencing (P < 0.001), as illustrated in Figure 3a. On IARS2 knockdown in Hela cells, there was a notable decrease in the proportion of EdU-positive nuclei, signifying the inhibitory effect on the proliferation of cervical cancer cells (P < 0.001) [Figure 3b and c]. Furthermore, the results from the Transwell assay provided compelling evidence that the in vitro migration of Hela cells was effectively suppressed following IARS2 silencing (P < 0.001) [Figure 3d and e].
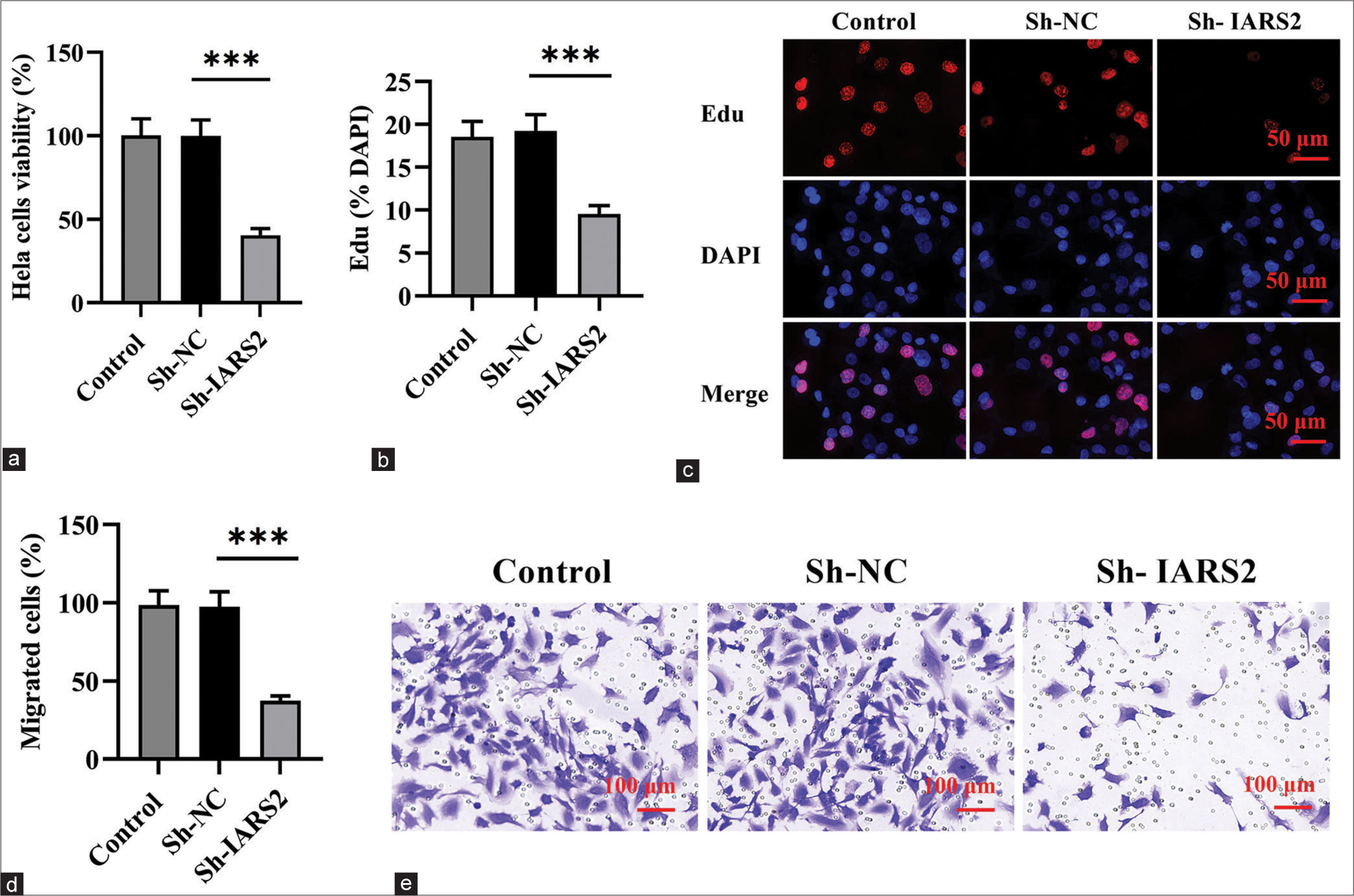
Export to PPT
The knockdown of IARS2 induces apoptosis in cervical cancer cellsSubsequently, we proceeded to explore the potential impact of IARS2 gene knockout on cell apoptosis. Within Hela cells, IARS2 silencing elicited a significant upregulation in the expression levels of cleaved caspase-3, and Bax proteins, concurrently with a reduction in the expression level of Bcl-2 protein (P < 0.001) [Figure 4a-d]. We observed a noteworthy increase in the proportion of terminal deoxynucleotidyl transferase dUTP nick end labeling-positive nuclei and the count of apoptotic cells in Hela cells subjected to IARS2 knockdown (P < 0.001) [Figure 4e and f]. Thus, IARS2 knockdown induces apoptosis in cervical cancer cells, as evidenced by these molecular and cellular alterations.
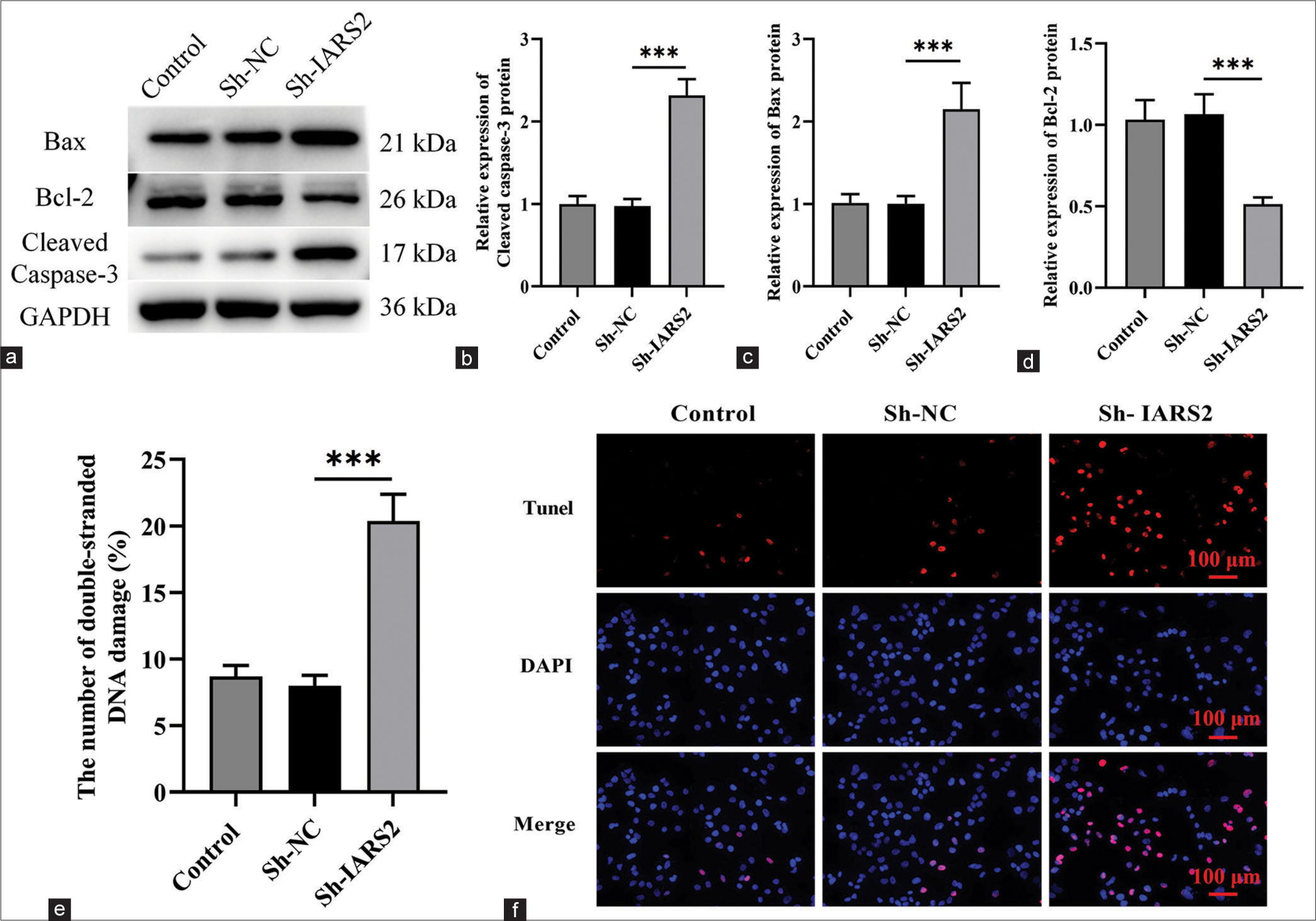
Export to PPT
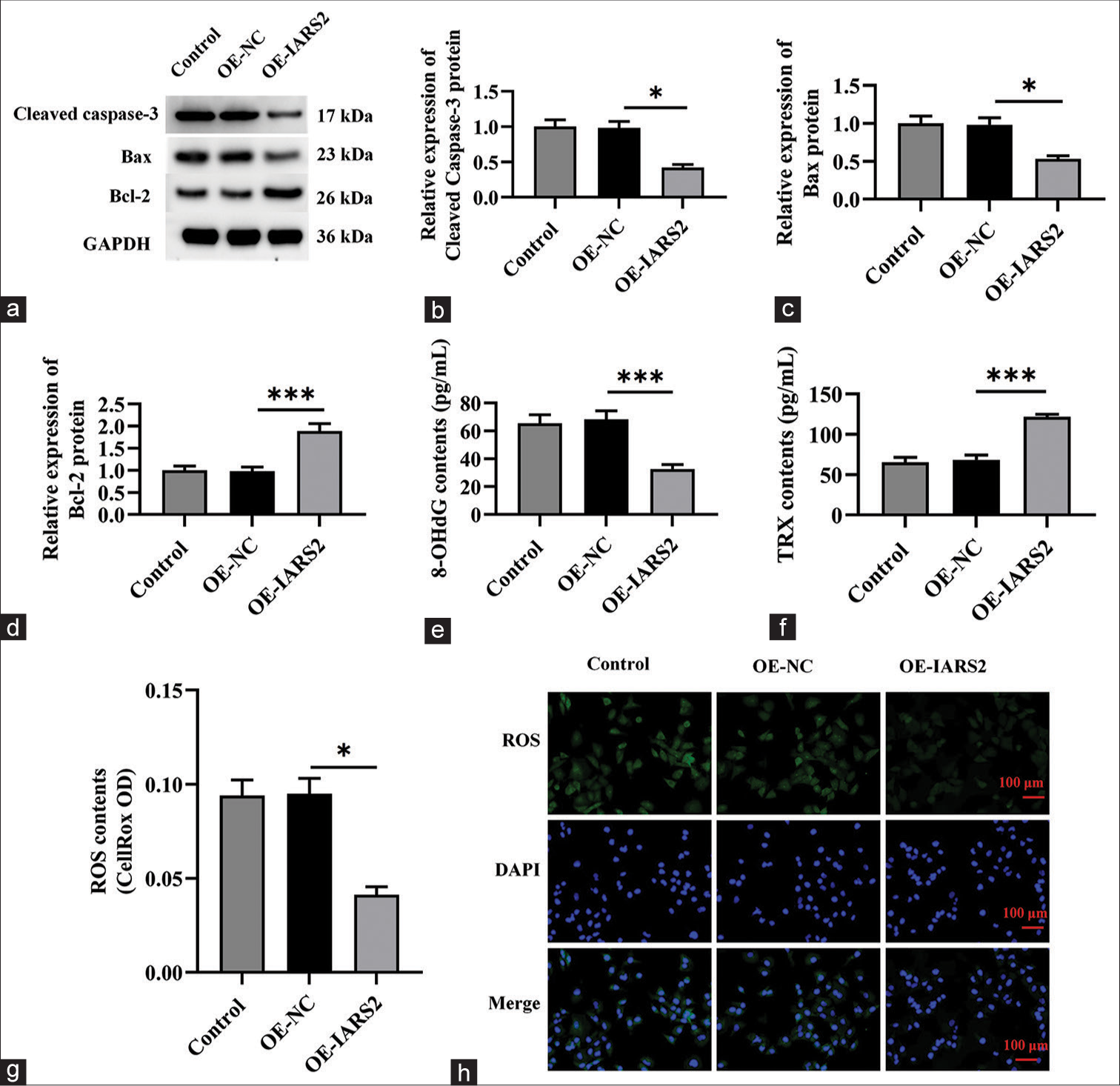
Overexpression of IARS2 induces pro-carcinogenic activity in primary cervical cancer cellsThe preceding findings suggest that the silencing of IARS2 confers robust anti-cancer activity in cervical cancer cells. Consequently, we postulated that the overexpression of IARS2 might lead to contrasting effects. To substantiate this hypothesis, we established an IARS2 overexpression cell model in Hela cells. As depicted in Figure 5a-c, the mRNA and protein levels of IARS2 in the OE-IARS2 group were markedly higher than those in the overexpression-negative control (OE-NC) group and control group (P < 0.001). Moreover, Hela cells with IARS2 overexpression exhibited an elevation in ATP levels (P < 0.05) [Figure 5d]. The overexpression of IARS2 facilitated proliferation in Hela cells, as evidenced by an augmented nuclear EdU incorporation (P < 0.05) [Figure 5e and f]. In addition, IARS2 overexpression expedited the in vitro migration of Hela cells (P < 0.05) [Figure 5g and h]. Furthermore, compared to the control group and OE-NC group, overexpression of IARS2 significantly reduced the protein expression of Cleaved caspase-3 and Bax, while increasing the protein expression of Bcl-2 (P < 0.05, and P < 0.001) [Figure 6a-d]. In addition, overexpression of IARS2 significantly decreased the levels of 8-Hydroxy-2'-deoxyguanosine and increased the level of Thioredoxin (P < 0.001) [Figure 6e and f]. Results from [Figure 6g and h] demonstrate that overexpression of IARS2 significantly reduced the fluorescence intensity of ROS staining (P < 0.05).
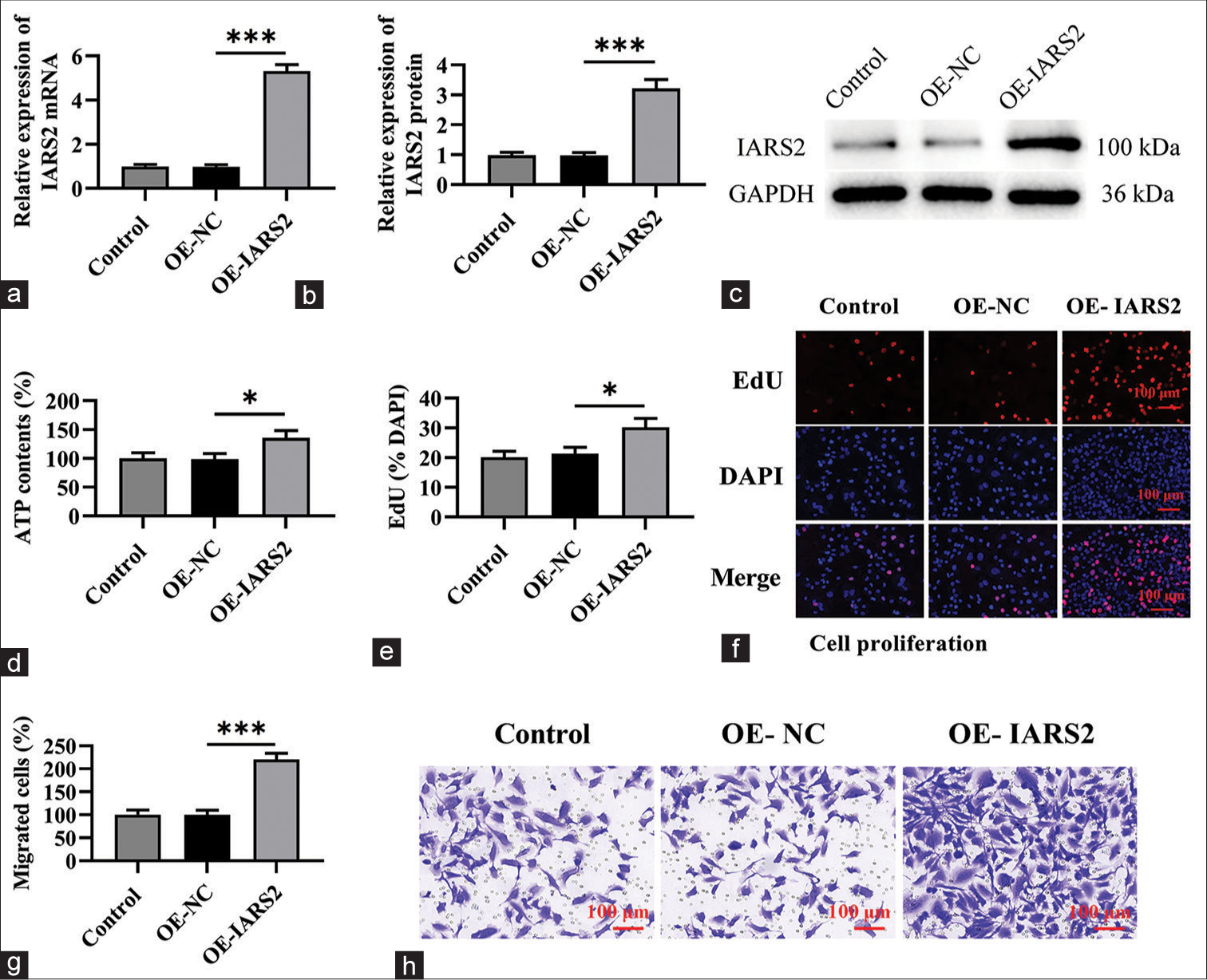
Export to PPT
Export to PPT
DISCUSSIONMitochondria play a pivotal role in the initiation and advancement of cervical cancer.[11,12] Elevated mitochondrial transcription mechanisms and subsequent protein translation are observed in diverse cancer types, such as breast cancer, liver cancer, cervical cancer, ovarian cancer, and numerous others. This heightened activity is associated with an unfavorable prognosis among cancer patients.[13-15] The primary function of IARS2 in cells is to encode isoleucyltRNA synthetase, a key enzyme crucial for maintaining normal mitochondrial function.[16-18] Recent investigations have revealed a connection between the onset of tumors in cervical cancer and a shift from ineffective metabolism to efficient mitochondrial metabolism.[18,19] Mitochondrial function is closely associated with cellular energy metabolism, and human papillomavirus (HPV) infection may affect the structure and function of mitochondria. This impact could potentially involve the proliferation and survival of cervical cancer cells.[20] Mitochondria serve as the primary site for cellular redox reactions and are also the primary location for oxidative stress occurrence within the cell.[21] Oxidative stress can potentially lead to DNA damage, cell apoptosis, and other processes, thereby promoting the occurrence of cancer.[22] In this scenario, oxidative damage may promote the occurrence of cancer by inducing processes such as DNA damage and apoptosis. DNA damage can result in impairment of the cell’s genetic material, potentially leading to mutations and the development of cancer. Liu et al.’s study suggests that depletion of KCNA1 can inhibit the growth, proliferation, metastasis of Hela cells by modulating the Hedgehog, Wnt, and Notch signaling pathways, leading to mitochondrial dysfunction.[23] Likewise, disruptions in mitochondrial function play a pivotal role in the onset and progression of prostate cancer.[24] Research suggests that both the quantity and activity of mitochondria are heightened in prostate cancer cells, potentially attributable to the uptake of lactate by cancer-associated fibroblasts.[25] Recent research findings suggest that the silencing of IARS2 impairs the mitochondrial function of cancer cells.[17] Recent studies have pointed out that IARS2 promotes the progression of non-small-cell lung cancer tumors by activating the Protein Kinase B (AKT)/mammalian target of rapamycin pathway.[26]
IARS2 broadly impacts mitochondrial function by maintaining mitochondrial protein synthesis, stabilizing mitochondrial membrane potential, supporting mitochondrial complex I function, and promoting ATP production.[26,27] Its high expression in cervical cancer cells aids in cell survival, proliferation, and migration, while its silencing leads to mitochondrial dysfunction, energy crisis, and cell apoptosis. IARS2 is considered crucial for maintaining mitochondrial activity and function. Therefore, when the IARS2 gene is knocked out, it may lead to damage to the structure and function of mitochondria, thereby affecting their normal function. Experimental results indicate that IARS2 knockout results in a decrease in mitochondrial membrane potential, possibly due to changes in mitochondrial inner membrane permeability or impairment in the activity of mitochondrial respiratory chain complexes. This decrease in mitochondrial membrane potential may further affect energy synthesis and ROS production in mitochondria. IARS2 knockout leads to elevated levels of oxidative stress, possibly due to increased ROS production resulting from impaired mitochondrial function. Excessive ROS production may cause oxidative damage within cells, leading to processes such as apoptosis and other cellular injuries. IARS2 knockout results in inhibition of the activity of mitochondrial complex I, further affecting energy metabolism and ATP production in mitochondria. This may lead to cells lacking sufficient energy to maintain normal biological functions. In summary, IARS2 gene knockout may lead to mitochondrial oxidative stress and damage by affecting mitochondrial structure and function, resulting in decreased mitochondrial membrane potential, increased ROS production, and impaired activity of mitochondrial complex I.
The findings of this study underscore the crucial role of the mitochondrial protein IARS2 in cervical cancer. The research reveals a significant elevation in both mRNA and protein levels of IARS2 in cervical cancer cells, particularly in Hela cells. Silencing the IARS2 gene using ShRNA virus significantly inhibits the vitality, proliferation, and migration of cervical cancer cells, concurrently inducing apoptosis. Conversely, overexpression of IARS2 enhances the proliferation and migration of cervical cancer cells. Hence, the overexpression of IARS2 holds significant implications for the growth of cervical cancer cells, establishing it as a novel diagnostic and therapeutic target.
Moreover, our study elucidates the critical role of IARS2 in preserving mitochondrial function in cervical cancer cells. In Hela cells, the application of IARS2 shRNA induces mitochondrial depolarization, promotes ROS generation, inhibits mitochondrial complex I, and results in ATP depletion. Conversely, overexpression of IARS2 results in an increase in ATP levels. Consequently, the loss of IARS2 impairs mitochondrial function and hinders the growth of cervical cancer cells.
CONCLUSIONTherefore, IARS2 emerges as a crucial mitochondrial protein for the growth of cervical cancer cells, establishing itself as a novel and promising therapeutic target.
AVAILABILITY OF DATA AND MATERIALSThe data that support the findings of this study are available from the corresponding author upon reasonable request.
ABBREVIATIONSIARS2- Isoleucyl-tRNA synthetase 2
ShRNA- Short Hairpin RNA
ATP- Adenosine triphosphate
ROS- Reactive oxygen specie
TUNEL staining- Terminal deoxynucleotidyl transferase dUTP nick end labeling staining
CCK-8- Cell counting kit-8
ELISA- Enzyme-linked immunosorbent assay
HE- Hematoxylin and Eosin staining
NC- Negative control
OE- Overexpression
GAPDH- Glyceralde-hyde-3-phosphate dehydrogenase
A- Adenosine
C- Cytosine
G- Guanine
T- Thymine.
AUTHOR CONTRIBUTIONSXM and NL: Interpreted the data and wrote the manuscript; BG: Designed the study; NL: Contributed to writing the manuscript. All authors read and approved the final version of the manuscript.
Comments (0)