The observable dynamics of individual neurons are currently well-understood at a biophysical level. However, there is still much to be gained from studying the behavior of large-scale brain networks. Specifically, it is not yet fully understood how the complex dynamics of these networks give rise to higher-order brain functions. Accordingly, simulations of these brain networks can provide new insights into brain workings and human behavior (Murray et al., 2018; Lam et al., 2022). Furthermore, it is believed that brain-network research can also lead to better understanding of treatments for psychiatric disorders (Murray et al., 2018; Einevoll et al., 2019). For a comprehensive understanding of the brain, information from multiple scales is required. Simulations of detailed neural models of (large regions of) the human brain, which comprise around 86 billion neurons and 1 trillion synapses, based on latest estimates, are nowadays recognized to call for exascale computing (Amunts and Lippert, 2021).
Present-day High-Performance Computing (HPC) solutions have already delivered impressive brain simulations, however, their limitations become manifest once the challenges of simulating life-sized brain models are identified:
1. Performance and scalability. The computational power of a single processing unit plays a vital role in a simulation's overall performance. Therefore, HPC accelerators, which pack substantial computational throughput, are essential brain-simulation components. However, performance efficiency is equally crucial to pure performance, since we know that a single processing unit (or accelerator) cannot suffice for simulating the whole brain. The solution is to enlist more processing units to the simulation effort. The speedup gained from adding more processing units correlates with a program's inherent parallelism. Gustafson's Law (Gustafson, 1988) stipulates that significant speedup can be achieved with additional computational resources the higher the parallelizable portion (0 ≤ p ≤ 1) of the program is. Figure 1 shows speedups when enlisting increasing numbers of processing units for p = 0.9 (solid blue line) and p = 0.5 (solid green line). But these are ideal speedups in the absence of overheads that can bring system performance (efficiency) down. Nowadays, the so-called Memory-Wall problem (McKee, 2004) has emerged as a main challenge limiting achievable speeds; that is, memory speeds cannot keep up with accelerator speeds, effectively constraining achievable performance. This can be seen in the respective dotted lines in Figure 1, which represent sub-linear performance scaling. To tackle this issue, it is crucial to minimize memory accesses and keep data as close to the processing unit as possible. Over networks of processing units, the problem becomes even more pronounced, especially in the case of simulating large-scale and realistic brain models that exhibit dense synaptic activity among their nuclei. That is why—besides enlisting powerful accelerators—it is imperative to also implement low-latency and high-throughput interconnects to ensure good performance scalability and efficient utilization of all computational resources, minimizing memory accesses (Ishii et al., 2017). This is exemplified in cases such as Pronold et al. (2022), where network communication dominates simulation time, and communication time deteriorates with enlisting more CPU MPI processes.
2. Flexibility. Simulator flexibility is a crucial property of modern-day simulation platforms since the computational-neuroscience community is in constant flux, always tweaking model aspects and tuning their parameters. Consequently, brain simulators require sufficient flexibility and modularity in order to cover a wide variety of configurations needed for research.
3. Usability. For all their complexity, mounting simulations should be as easy to use as possible for neuroscientists. What is more, if an HPC simulator should make use of hardware accelerators to improve its performance, as is the current trend, then harnessing its full potential should–ideally–not require specialized knowledge from an acceleration expert working next to the neuromodeler.
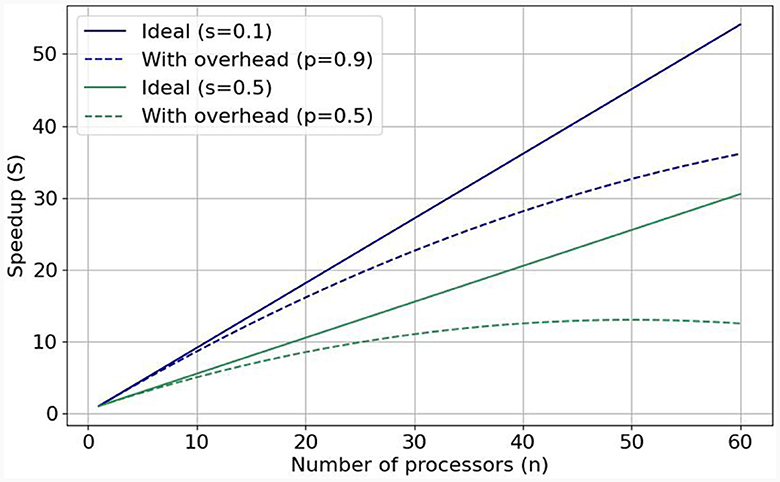
Figure 1. Visualization of Gustafson's law and weak scaling: with predicted (dotted lines, realistic) and without (solid lines, ideal) added communication overhead when the parallelizable fraction is equal to 0.9 and to 0.5.
In this work, we advocate the use of exascale-ready computing methods for facilitating the steep requirements of large-scale brain simulations. Traditional HPC solutions are known to fall short of meeting these requirements. To eschew the inherent memory bottleneck of conventional (von-Neumann) processing technologies such as Central Processing Units (CPUs) and Graphics Processing Units (GPUs), Field-Programmable Gate Array (FPGA) acceleration is recognized as one of the most robust platforms for attaining scalable performance when discounting exotic approaches such as quantum computing. Until now, their low usability (which is neurosimulator challenge 3) has been the main deterrent neuromodelers consistently adopting them in the field. In this work, we will demonstrate that this final barrier can be largely overcome through the combination of key enabling technologies and special design methods.
In terms of methods, firstly, the use of modern FPGAs allows designing dataflow-computing kernels instead of following the typical control-flow (i.e., von-Neumann) approach, which results in significant performance gains for data-intensive workloads, such as brain simulation (Flynn et al., 2013). Building on top of this dataflow-computing paradigm, parameterizable FPGA libraries such as flexHH (Miedema, 2019) have been proposed for simulating a gamut of biologically plausible brain models. The trivial control flow of dataflow kernels, in turn, permits the design of very simplified hardware interconnects across FPGA accelerators, effectively leading to communication-efficient, systolic-array-like multi-FPGA ensembles. Thus, on the technology front, some dataflow-enabled FPGA platforms can offer the option for direct communication links between them, allowing for direct, low-latency, and high-throughput connections, without the interference of a host CPU. This makes them highly promising for exhibiting good performance scalability (Pell et al., 2013). Finally, the addition of latest High-Bandwidth Memory (HBM) modules on FPGA chips significantly improves their performance for memory-bound applications (Wang et al., 2020).
The aforementioned aspects point to specific multi-FPGA platforms as a highly promising candidate for tackling the three identified challenges for biologically detailed, exascale-ready, large-scale brain simulations. In this article, we present ExaFlexHH, a dataflow-based, performance-scalable, and user-friendly brain simulation library. Our contributions are as follows:
• A concrete proposal for attaining exascale-ready brain simulations based on an ensemble of cutting-edge technologies and methods.
• A review and taxonomy of multi-accelerator platforms for brain simulations.
• A future-proof, scalable, multi-FPGA simulation library called ExaFlexHH, for eHH models that is synthesis-free, parameterizable, and flexible.
• A detailed performance evaluation of ExaFlexHH, including a performance model for making future, scale-out projections.
The remainder of this paper is organized as follows: Related works are presented in Section 2. In Section 3, we provide crucial background information and detail the ExaFlexHH implementation. In Section 4, we present our evaluation results. Section 5 examines performance bottlenecks and evaluates the potential for improvement if some of these bottlenecks are addressed. Finally, in Section 6, we present our conclusions.
2 Related workMany HPC works have aimed at brain research in recent years. We have chosen only works that meet the following criteria: firstly, works that utilize high-performance accelerators since these are a crucial component in achieving the large-scale and highly scalable simulations required for brain research. Secondly, works that utilize multi-accelerator computation, since this is essential for achieving the required performance density for large-scale and highly detailed brain simulations. We have, thus, excluded SpiNNaker and NEST (Gewaltig and Diesmann, 2007) due to the absence of accelerator support as well as Brain2 (Stimberg et al., 2019) and GeNN (Yavuz et al., 2016) due to a lack of support for scale-out acceleration. Finally, platforms focused on machine learning and cognitive neuroscience are excluded as generally unsuitable for simulating biologically detailed models; therefore, BiCoSS (Yang et al., 2021), Loihi (Davies et al., 2018), and Tianjic (Deng et al., 2020) have been dropped.
An overview of eligible works is shown in Table 1. We have organized information into three main categories, largely matching the three challenges set in the previous section, as follows: (I) Performance (scalability): The number of accelerators per node, the number of nodes, and the type of connections used will be specified and presented. This information will give insight into the potential performance that can be achieved and the cost of utilizing the system. (II) User experience: Computational-neuroscience language support such as PyNN (Davison et al., 2009) and NeuroML (Cannon et al., 2014). Also, advanced and easy-to-use graphical user interfaces (GUIs) are contributing to user experience. We distinguish three levels of flexibility: no ( ), partial (+) and full (++) flexibility. (III) Biological plausibility: The neuron-model(s) and type(s) of synapses are specified here. Compared to other types of models, HH and particularly eHH models incur high computational costs (Izhikevich, 2004; Kozloski and Wagner, 2011), but more crucially, also high communication costs due to the detailed modeling of electrical and chemical synaptic activity. Of the two, chemical-synapse activations are relatively slow events and can be simulated in an event-based manner, reducing communication costs. In contrast, electrical synapses (i.e., gap junctions) require continuous interneuron communication, stressing multinode-accelerator data transfers, which can limit throughput and latency (Hahne et al., 2015; Jordan et al., 2018).
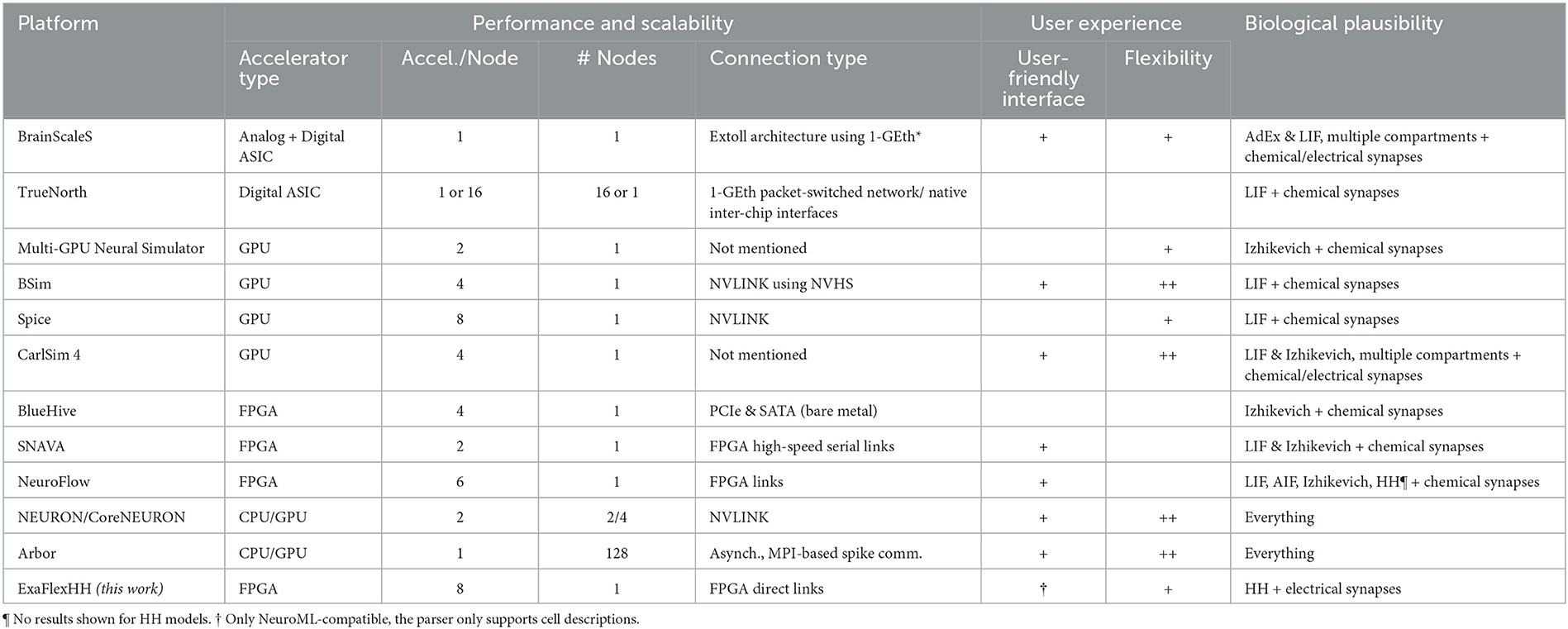
Table 1. Comparison of ExaFlexHH with other brain simulation developments.
BrainScaleS-2 (Pehle et al., 2022), is an analog hardware platform for emulating spiking neural networks. It supports Adaptive-Exponential Integrate-and-Fire (AdEx) or integrate-and-fire (I&F) neurons and allows for multi-compartmental features (Kaiser et al., 2022), and conductance-based synapses. The platform utilizes digital chips for control and plasticity management and the EXTOLL network protocol (Neuwirth et al., 2015) in combination with FPGAs for interconnectivity. BrainScaleS-2 can be interfaced with through PyNN (Müller et al., 2022) providing a user-friendly interface. However, the platform has not yet been tested for performance scalability and consequently this remains uncertain. TrueNorth (Akopyan et al., 2015) is a specialized chip for neural simulations that has been demonstrated in different configurations such as a single chip on a NS1e board, 16 NS1e boards connected through a 1 GbE packet-switch network, and the NS16e platform on a 4 × 4 board. However, its performance scalability has not been evaluated making good scalability uncertain. Additionally, TrueNorth chips only support I&F models, limiting their biological plausibility. The Corelet developer kit (Amir et al., 2013) requires learning its environment, instead of using standardized languages such as NeuroML or PyNN. Both aforementioned platforms are Application-Specific Integrated Circuits (ASICs). ASIC solutions offer excellent performance, scalability and energy efficiency but lack the level of flexibility needed in the constantly evolving field of computational neuroscience. Even trivial model changes can easily result in a new development cycle, significantly delaying the research process and stacking costs. Consequently, such solutions do not meet the general brain-simulation needs.
In Thibeault et al. (2011), Qu et al. (2020), and Bautembach et al. (2021), three GPU-based simulation platforms for Spiking Neural Networks (SNNs) are introduced: a multi-GPU neural (mGPUns) simulator, BSim, and Spice. The scalability results of mGPUns are constrained as only the results of 1 and 2 GPUs are shown and thus insight is limited. Both the performance results of BSim and Spice have shown limited performance scalability, ranging from as low as 1.6 comparing 4-GPU execution to single-GPU with BSIM execution to ≈5.1 when using 8 GPUs compared to 1 GPU with Spice. Additionally, the simulators lack the support of HH-type models and only BSim supports PyNN. In contrast, CARLsim 4 (Chou et al., 2018) is a multi-GPU simulator that aims at supporting a wide range of neural models and synapse types including Izhikevich models, multiple compartments, and current and conductance-based synapses with plasticity. CARLsim 4 offers tools for parameter tuning and visualization and uses a custom API rather than standard languages like NeuroML or PyNN. However, performance evaluations show limited performance scalability as the maximum increase in performance is 1.95 × and 2.44 × when using 2 and 4 GPUs, respectively, compared to 1 GPU; also, there is no support for eHH models. Consequently, it is not optimally qualified for large-scale, biologically detailed simulations. Overall, all discussed GPU platforms exhibit excellent performance, flexibility, and usability. However, they are von-Neumann architectures with all that this entails, including thread synchronization, instruction overheads and memory latencies (Hameed et al., 2010; Yazdanpanah et al., 2013). These limitations become especially evident when energy efficiency is a consideration (Lant et al., 2019). While there have been promising developments such as NVLink, performance scalability is questionable due to difficulties in efficiently exploiting such developments (Li et al., 2019). Therefore, the GPU solutions are not considered optimal to achieve ideal performance scalability.
In the field of FPGAs, Bluehive (Moore et al., 2012) is a computing platform that utilizes 16 FPGAs in a rack, connected via PCIe and a custom PCIe-to-SATA adapter for a reconfigurable topology. It can simulate 64k Izhikivich neurons with 64 million synapses per FPGA in real-time on a four FPGA setup. However, the system does not provide any scalability results. Additionally, Bluehive is limited to Izhikevich models with no easy to add new functionalities and has a lack of a user-friendly interface reducing the accessibility for non-experts. SNAVA (Sripad et al., 2018) is an FPGA-based neural simulation platform with a custom interface for model selection and connectivity. While it can simulate leaky integrate-and-fire (LIF) and Izhikevich neurons, it lacks support for HH-type models and widely used languages like PyNN and NeuroML. Using fixed-point simulation for performance may impact accuracy in stiffer, biophysical models like eHH. Although it is designed for scalability with an expandable ring structure, experimental results are limited to a two-FPGA network, and model updates' impact on synthesis cycles introduces uncertainty and challenges SNAVA's flexibility. NeuroFlow (Cheung et al., 2016) is another multi-FPGA neural simulation platform with PyNN compatibility. It supports a range of models including HH neurons. This makes NeuroFlow one of the most user-friendly and complete neural simulation platforms available. However, it does not support gap-junction connectivity and multi-compartment models. Additionally, performance and scalability results are limited to the simpler Izhikevich models and event-driven implementations, with synapses between neurons on the same or neighboring FPGAs. Therefore, while NeuroFlow is promising, its performance and scalability for complex cases remain to be seen. In all FPGA solutions, the hardware is configured specifically for each application, delivering high performance, while also providing higher levels of (re)modeling flexibility as well as energy efficiency (Chow et al., 2012; Guo et al., 2012; Arram et al., 2013; Gan et al., 2013). Unfortunately, flexibility comes at the cost of notoriously low levels of programming ease compared to GPUs due to the stringent hardware-programming languages involved (e.g., VHDL, Verilog) as well as the large, hardware-synthesis debug cycles.
Finally, we include two full-fledged neurosimulator environments, which support multinode simulations using CPU-only or a mix of CPU & GPU implementations. The community standard simulator NEURON (Hines and Carnevale, 1997) integrates HPC solutions through CoreNEURON (Kumbhar et al., 2019; Awile et al., 2022). Therefore, it supports simulations on multi-threaded CPUs and GPUs, and multi-node processing through the use of MPI. This brings significant performance benefits to NEURON. However, its performance scalability is still far from ideal as it is constrained by the previously mentioned von-Neumann limitations. Arbor (Abi Akar et al., 2019) is a neural simulator focusing on high-performance processing and multi-compartmental neuron models, including eHH with gap junctions. Additionally, Arbor is designed to be user-friendly, providing an object-oriented interface. However, its performance scalability when modeling complex connectivity is unclear, and centralized spike exchange between neurons may limit its ability to scale efficiently. Furthermore, the capability to handle large-scale simulations with gap-junction connectivity is not demonstrated.
ExaFlexHH is a high-performance, hardware library for simulating biologically plausible eHH models on one or multiple FPGAs. The use of the dataflow paradigm allows for efficient utilization of hardware acceleration and support for multiple FPGAs connected within a single node in a ring structure allows for low-latency interconnects. The system's modular design allows easy modification of parameters without re-synthesis, while NeuroML compliance ensures user-friendliness. Though communication can extend seamlessly outside a single compute node, ExaFlexHH has been currently demonstrated on as many as 8 FPGAs on a single compute node, leaving multi-node as future work. Despite this limitation, ExaFlexHH provides a flexible, highly scalable, and high-performance option for the simulation of large-scale eHH models.
3 MethodThis section begins with the discussion of HH-type models and our use case, the Inferior Olive, and an explanation of why this model is suitable as a benchmark in Section 3.1. The subsequent Section 3.2 discusses the Maxeler system and the dataflow paradigm. Then, in Section 3.3 the predecessor of ExaFlexHH, flexHH is discussed. Finally, the implementation is detailed in Section 3.4.
3.1 Hodgkin-Huxley-type modelsThe HH neural networks described here are represented by a set of Ordinary Differential Equations (ODEs). Therefore, an ODE solver is required to solve (i.e., simulate) these models. The simplest ODE solver is the forward-Euler, shown in Equation (1). Here, un represents the approximated state variables step n, Δt denotes the time-step size, and f a describes the vector of state derivatives. This process progresses iteratively for a simulation.
un+1=un+Δt·f(un) (1)In HH-type models, two types of state variables are involved: membrane voltages across cellular compartments, and gate variables indicating ion-channel openings. The voltage derivative for a specific compartment i in an HH-type model is computed as per Equation (2), where C signifies membrane capacitance, Iapp, i is the applied current representing external stimuli, Ichannels, i aggregates all ion-channel currents, Ileak, i indicates leakage current, and Imc, i and Igap, i reflect currents from inter-compartment connections and gap junctions, respectively. Notably, these latter terms are model-dependent and may be excluded if not applicable. For example, the original HH only consists of a single cell and a single compartment and therefore, does not include Imc, i and Igap, i. A network of 3 compartmental neurons is given in Figure 2. This figure shows three compartments, the currents per compartment, and all-to-all, through gap junctions, connected network of 8 neurons.
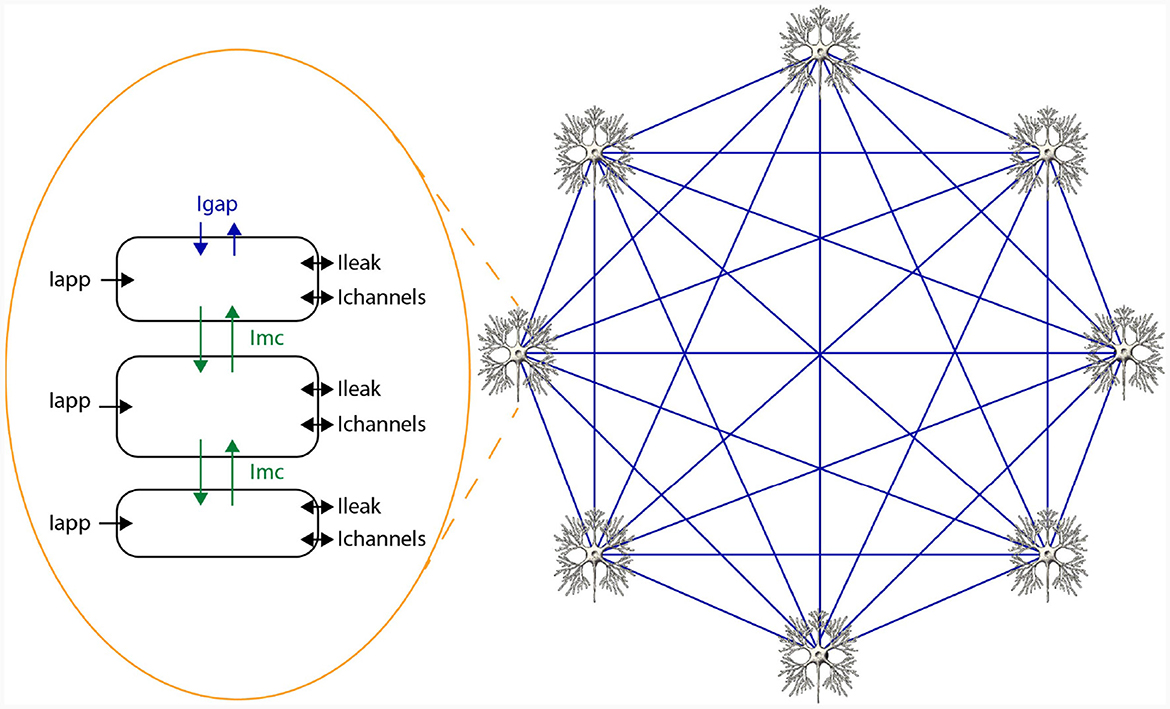
Figure 2. Schematic overview of a network of HH-type neurons.
Iapp, i can be defined by any arbitrary function, while Ichannels, i follows Equation (3). In this equation, Nchannels, i is the number of channels for compartment i, gc, j is the conductance, and Vc, j is the reverse voltage of channel j. Furthermore, Ichannels, i involves yProdj, the product of gate activation variables of channel j calculated using Equation (4). In this equation Mgates[j] represents the amount of different gate types and pk is an integer that counts the number of gates for a given type within the channel.
dVidt=Iapp,i−Ichannels,i−Ileak,i− Imc,i− Igap,iC (2) Ichannels,i=∑j=0Nchannels,i−1Ichannel,j =∑j=0Nchannels−1gc,j(V−Vc,j)yProdj (3) yProdj=∏k=0Mgates[j]-1ykpk (4)For models supporting multiple compartments, Imc, i is added to represent the current between adjacent compartments. To calculate the current flowing between compartments, we use a similar equation as in Schweighofer et al. (1999), shown in Equation (5). This equation incorporates the number of linked compartments Ncomps, i to compartment i, the internal conductance gint, the surface ratio of adjoining compartments pi, j, and their respective membrane voltages (Vi, Vj).
Imc,i=gint∑j=0Ncomps,i-1Vi-Vjpi,j (5)Gap junctions, the inter-cellular connections, are modeled following a generalized approach from Schweighofer et al. (2004) by calculating Igap, i through Equation (6), where Vi, j is the voltage difference between cell i and j, c0, c1, and c2 are identical between connections, and wi, j represents the weight between compartments i and j, where j belongs to a different cell than i, therefore enhancing data efficiency and model adaptability.
Igap,i=∑j=0Ncells-1(wi,j(c0exp(c1·Vi,j2)+c2)Vi,j) (6)The derivatives of the gate-activation variables represented by yj are also required and can be determined via Equation (7) and/or Equation (8). These involve transition rates αj and βj or the target value infj and the time constant τj, which are generally determined by exponential functions. For reference, the derivatives of the gate-activation variables of the original HH model (Hodgkin and Huxley, 1952) are presented in Equations (9–17).
dyjdt=(1-yj)·αj-yj·βj (7) dyjdt=infj-yjτj (8) dndt=αn(1-n)-βnn (9) dmdt=αm(1-m)-βmm (10) dhdt=αh(1-h)-βhh (11) αn=0.01(V+10)exp(V+1010)-1 (12) βn=0.125exp(V/80) (13) αm=0.1(V+25)exp(V+2510)-1 (14) βm=4exp(V18) (15) αh=0.07exp(V20) (16) βh=1exp(V+3010)+1 (17)The model used to benchmark ExaFlexHH is a model of the Inferior Olive (IO) which is a brain region implicated in learning and online motor control (Schweighofer et al., 2013). De Gruijl et al. (2012) developed a model of an IO network employing an eHH description. The extensions incorporated in this model include more sophisticated ion gates, multiple compartments, and gap junctions. Specifically, each IO cell in the model consists of three compartments: the dendrite, soma, and axon. The sophisticated ion gates and the multi-compartmental structure augment the complexity as well as the computational demands belonging to the intracellular dynamics. Moreover, the inclusion of gap junctions among the dendrites, which represents instant, continuous interneuron connections, further intensifies the complexity by requiring communication among cells, therefore disrupting parallelism and posing a challenge to straightforward performance scaling. Given the biological plausibility, complexity, and computational requirements of this model, it is a fitting scenario for evaluating the ExaFlexHH framework. The equations used for the IO model are provided as Supplementary material. For an exhaustive description of the model, readers are directed to De Gruijl et al. (2012).
3.2 Maxeler system and dataflow paradigmNeuron dynamical equations typically require minimal control, such as a few if/else statements, making them well-suited for the dataflow paradigm. This paradigm, especially when implemented using FPGAs, can be efficiently leveraged. Maxeler Technologies offers a unique solution in this space with its Data-Flow Engines (DFEs) and associated tools (Pell et al., 2013). The DFEs are FPGA-based hardware that are programmed via the use of Maxeler tools and excel in exploiting the dataflow paradigm.
In the dataflow paradigm, traditional control logic is mostly absent. Compute dependencies are resolved statically, at compile time. Control is effectively reduced to counters that advance data through execution units in the datapath. This approach allows for the majority of FPGA resources to be dedicated to computation rather than control logic. Moreover, it enables implementation in a deeply pipelined manner, significantly enhancing computational throughput.
A key factor for efficient dataflow implementation on FPGA-based hardware is the use of on-chip memory. Contemporary FPGAs, such as the Xilinx Ultrascale+ (AMD, 2023), feature three levels of on-chip memory. The first level, utilizing logic slices and lookup tables, creates flexible RAM but is not efficient for larger memories. The second level, Block Random-Access Memory (BRAM), comprises physical memory units with up to 36 Kbits storage capacity. These units can be combined for greater capacity. The third level, UltraRAM (URAM), offers the largest storage (288 Kbits) but is the least flexible. The Maxeler tools abstract these memories and collectively call them FMem (Fast Memory). With the use of the Maxeler tools, the on-chip memory is classified as FMem. Additionally, the DFEs contain on-board DRAM, ranging in the order of tens of gigabytes, which is referred to as LMem (Large Memory) in the Maxeler tools.
Another advantage of using the dataflow paradigm with DFEs is scalability. DFEs connect directly via the MaxRing, which is a low-overhead, wired Daisy-chain connection among DFEs in a server node and accommodated via using unused PCIe pins on the mainboard. The MaxRing, thus, offers high-bandwidth, low-latency, and highly scalable interconnects. This facilitates deeper pipeline designs, increased parallelism, and a highly scalable platform architecture that ExaFlexHH intends to harness.
Programming the Maxeler system involves three fundamental parts:
1. CPU-Host Code: Written in C, this code initializes data, coordinates DFEs, and manages I/O (Input/Output) between the CPU-host and DFEs.
2. Kernel Code: Using MaxJ, an extended version of Java, this code defines the functionality of the kernel(s) on the DFEs.
3. Manager: Also defined in MaxJ, the manager configures I/O for the kernels, including on-board DRAM, inter-kernel communication, MaxRing, and CPU-host interactions. It also sets hardware-specific configurations like frequency and synthesis strategies.
The toolflow process begins with the MaxCompiler translating MaxJ kernel code into a dataflow graph. The MaxCompiler uses this graph and the manager description to generate VHDL code. This code is then utilized by FPGA vendor tools for implementation processes like synthesis and place-and-route, ultimately producing a bitfile for use with the C code on the CPU-host. This process is visualized in Figure 3. In this figure, it can be seen that the lines of code are directly translated to functional units on the DFE. Additionally, the figure depicts how multiple DFEs can be connected through MaxRing to construct a larger dataflow graph. Importantly, this graph features a pipelined architecture, thereby significantly enhancing parallelism.
Comments (0)